Get Better Protease Detection With A High-Sensitivity Protease Detection Assay
See how to improve your ability to detect and quantify protease activity accurately with a high-sensitivity protease detection assay – the Red Protease Detection Kit.
Section Overview
What Are Proteases?
Proteases, also known as proteinases or proteolytic enzymes, are a large group of enzymes that catalyze proteolysis. This proteolysis occurs by hydrolysis (reaction that occurs as water breaks bonds) of proteins into smaller polypeptides or single amino acids.1,2,3
By breaking down proteins, proteases are involved in the control of many key physiological processes such as cell-cycle progression, cell proliferation and cell death, DNA replication, coagulation, wound healing, and the immune response.4 Proteases have a specific region called the active site where catalysis occurs. This site typically contains amino acid residues that act as catalysts, facilitating the cleavage of peptide bonds. In most proteases, catalysis involves a nucleophilic attack on the carbonyl carbon of the peptide bond. The nucleophile is usually a side chain from an amino acid residue within the active site of the protease.5
Categories of Proteases
Proteases are categorized into six proteolytic mechanism groups:
- Serine proteases are enzymes that cleave peptide bonds in proteins, with serine serving as the nucleophilic amino acid at their active site.6
- Threonine proteases belong to a family of proteolytic enzymes characterized by a threonine residue within their active site. These residues play a crucial role in the catalytic subunits of the proteasome, and threonine proteases are activated by primary amines.7
- Cysteine proteases are enzymes involved in protein degradation, utilizing a common catalytic mechanism where a nucleophilic cysteine thiol participates in a catalytic dyad or triad assembly.8
- Aspartic proteases use two conserved aspartic acid residues in their active site to catalyze the cleavage of peptide substrates. They are most active under acidic pH conditions and employ an activated water molecule bound to aspartate residues for substrate catalysis. Almost all aspartic proteases are inhibited by pepstatin and do not form a covalent intermediate during cleavage.9
- Metalloproteases are a family of proteases that feature a metal ion at their active site, which acts as a catalyst in the hydrolysis of peptide bonds. The metal ion within each enzyme's core is crucial for facilitating specific enzymatic reactions, with zinc ions being particularly common in metalloproteases.10
- Glutamic acid proteases constitute a class of proteolytic enzymes that include a glutamic acid residue in their active site, contributing to their catalytic function.11
An alternative classification is based on its optimal active working pH and temperature dependence because the activity of proteases is influenced by pH and temperature. Different proteases have optimal pH ranges where they exhibit maximal catalytic activity. For instance, pepsin, an aspartic protease found in the stomach, is most active at acidic pH, while trypsin and chymotrypsin, serine proteases found in the small intestine, are active at neutral to slightly basic pH.12
Protease Function
The main function of proteases is in the digestion of long proteins chains into shorter fragments by cleaving the peptide bonds. The digestion can occur in an exopeptidase way (cleavage of the N- or C-terminal peptides bond) as aminopeptidases, carboxypeptidase A; or an endopeptidase way (such as trypsin, chymotrypsin, pepsin, papain, and elastase), breaking internal peptide bonds.13
How Proteases Work
The catalytic mechanism of proteases typically involves: Activation, Cleavage, and Product Release. Activation is the binding of the substrate to the enzyme, often inducing conformational changes that align the reactive groups. Cleavage is the nucleophilic attack on the peptide bond, leading to the formation of a tetrahedral intermediate resulting in the formation of two separate peptide products- product release.5
Proteases can be inhibited by specific inhibitors that target their active site residues or cofactors. For example, serine proteases are often inhibited by serine protease inhibitors like phenylmethylsulfonyl fluoride (PMSF), while aspartic proteases are inhibited by molecules like pepstatin.14
Understanding the mechanisms of protease catalysis is essential not only for biological processes but also for developing therapeutic agents that can selectively inhibit or activate proteases in various disease contexts.
High-Sensitivity Protease Detection
The Red Protease Detection Kit is a high-sensitivity protease detection assay designed for testing the presence of proteases, to screen protease inhibitors as potential drug molecules, as well as evaluation of general protease contamination in various samples.
The Red Protease Detection Kit provides a simple and rapid method for protease detection. In this kit, proteolysis is measured using 5 (6)-tetramethylrhodamine (TAMRA)-labeled casein as a substrate. When the TAMRA-labeled casein substrate is intact, its fluorescence is almost totally quenched. Upon proteolytic cleavage, fluorescence of the TAMRA-labeled peptide fragments increases proportionally to protease activity in the sample (Figure 1). The fluorescence of TAMRA-labeled peptide fragments is measured at λex / λem=535 nm / 595 nm.
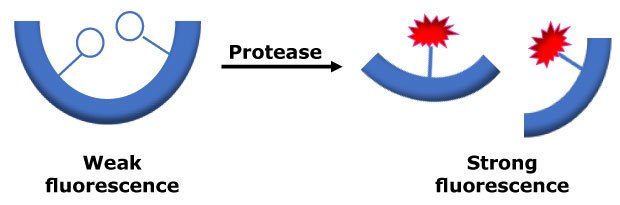
Figure 1.Schematic diagram of the proteolytic cleavage.
Another commonly used reagent required before separating the protein substrate and amino acid degradation products is Trichloroacetic Acid (TCA) precipitation. The TCA outcome is used during hazardous-material, low-throughput, and multiple-steps protocol (incubation, centrifugation). The TAMRA-labeled casein substrate is timesaving since no separation steps are required.
In addition, the TAMRA-labeled casein substrate has a pH-independent fluorescence. This makes the substrate applicable for a wide range active proteases and pH range.
This kit is designed for a 96-well plate format, and it can be easily calibrated to different sample types and to evaluate many samples in parallel.
Trypsin is provided in the kit as a reference protease for calibration and as a general protease control. Herein we show detection of Trypsin activity, in several time lengths and protease concentrations - Figures 2 and 3.
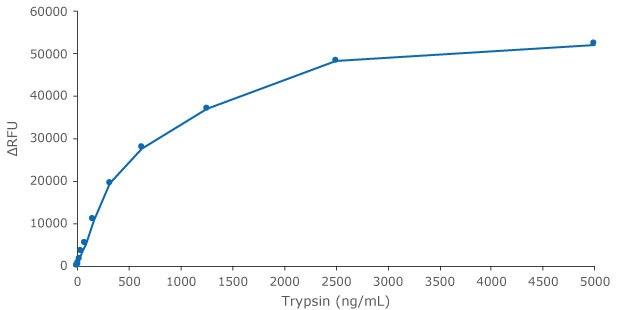
Figure 2.Trypsin standard curve preparation with the Red Protease Detection Kit showing trypsin activity after 1 hour of incubation. The fluorescence signal was measured with a filter fluorometer at λex / λem = 535 nm / 595 nm.
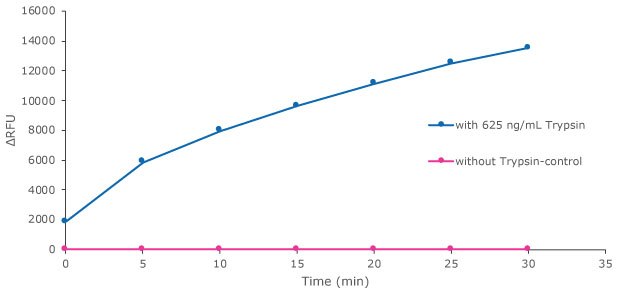
Figure 3.Trypsin protease activity was analyzed by Red Protease Detection Kit showing substrate proteolysis with trypsin. Substrate was incubated either with 625 ng/mL Trypsin, or without Trypsin as a control. The fluorescence signal was measured with a filter fluorometer at λex / λem = 535 nm / 595 nm.
The Protease Assay Buffer × 2 (pH ~7.5) provided in the kit is suitable mainly for proteases in the physiological pH range 4-12 (e.g., chymotrypsin and proteinase K). However, if activation compounds or other pH are required, a different specific buffer should be separately prepared.
This novel kit can be used for detection of many other proteases in their natural activity conditions, as is shown in Figure 4, pepsin protease activity is seen at pH of 2 and papain protease activity is seen at pH of 6.5.
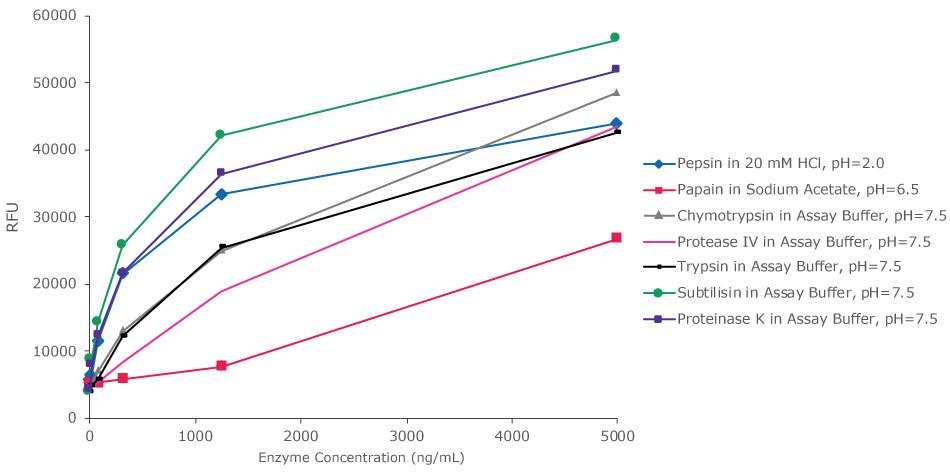
Figure 4.Substrate proteolysis by different proteases at different buffers after 30 minutes. Red Protease Detection Kit enables detection of varied proteases in a wide pH range. Activity at different protease concentrations was analyzed in their natural activity conditions or in Assay Buffer, pH ~7.5. The fluorescence signal was measured with a filter fluorometer at λex / λem = 535 nm / 595 nm.
The Red Protease Detection Kit enables better detection capabilities by minimizing background noise relative to the signal from the proteases, which improves the ability to detect and quantify protease activity accurately. Thereby, supports fast and easy detection of proteases in each sample. It serves as a high-throughput detection assay capable of analyzing multiple samples simultaneously and detecting the activity of numerous proteases. Experimental results probing the signal-to-noise ratio using the Red Protease Detection Kit are shown in Table 1, where trypsin is used at increasing concentrations to demonstrate an increase in signal-to-noise ratio as protease concentration increases.
Related Products
References
如要继续阅读,请登录或创建帐户。
暂无帐户?