Tag Removal Proteases for Recombinant Protein Purification
A reliable and robust protein purification method involves many considerations, including choosing the right columns for chromatography, calibrating the optimal binding and elution conditions, and the sequence of purification steps. Recombinant protein expression is a commonly used option when a pure protein in large quantities is desired. One benefit of recombinant protein expression is the ability to modify the gene encoding the protein to increase protein solubility and ease of purification via column purification methods. Commonly modified sequences, or tags include 6xHIS-tag and FLAG®-tag, or the addition of GST, MBP, and SUMO domains. The addition of biotin and biotinylated substances to the protein of interest is also commonly used as it’s strong, noncovalent binding to a streptavidin column makes it a suitable choice to obtain proteins of high purity. Explore our offering of recommended biotin protein purification reagents in the product selection table below.
Biotinylated proteases for on-column cleavage protein purification
Oftentimes the purification tag or carrier protein needs to be removed as it may interfere with the activity of the purified protein. Furthermore, various industries adhere to strict regulatory guidelines that often require tag-free proteins and tag removal may be required for downstream purification. To facilitate the proteolytic separation of the tag and protein of interest during purification, one of several protease recognition sequences is often introduced between the purification tag or the carrier domain and the protein (Figure 1).
Importantly, these tag-removal proteases must be specific and have a recognition sequence that will not appear randomly in the protein of interest. Commonly used proteases include TEV, HRV-3C, thrombin, and SUMO (Table 1). SUMO protease differs in that it does not recognize a small recognition sequence, but rather the actual SUMO domain secondary structure and cleaves the complete SUMO domain without leaving any additional amino acids to the protein of interest. As a result, SUMO protease is usually the best solution when an authentic N-terminal sequence is required.

Figure 1.Example of a fusion protein purification strategy using a protease for tag-removal from the protein of interest and subsequent analysis by SDS-PAGE.
Protein purification workflows that include tag removal often require that the tagged protein be first eluted from the affinity resin. To facilitate tag removal, protease is added to cleave the tag and an additional purification step is performed to separate the removed tag and remaining protease from the protein of interest. An alternative strategy is to cleave the protein of interest from the fusion tag while it is bound to the affinity column. This strategy is advantageous as it tends to improve the purity level of the eluted protein, eliminates the need for additional steps, and allows flexibility in the composition of the elution buffer. However, this strategy may be challenging as the tag-removal protease carries the same purification tag as the protein of interest, resulting in the binding of the tag-removal protease to the affinity resin which sterically limits its proteolytic activity.
Biotin protein purification
Providing a solution to the previously mentioned challenge of limited proteolytic activity, we now offer versions of the most popular tag removal proteases. Our proteases are free of common purification tags, including His-tag, FLAG®-tag reagents, GST & MBP so that these proteases will not bind to any common affinity purification resin, allowing for maximal tag removal activity. As the typical amount of protease added is less than 1% of the eluted protein, it might not be necessary to remove the tag-removal protease at all. However, several of our proteases are also biotinylated if tag removal is required, allowing for fast and easy removal of the protease using any Avidin or Streptavidin resin (Figure 2 and Table 1).

Figure 2.The on-column tag-removal process eliminates most impurities that co-bind to affinity resins, including metal binding proteins on Ni-chelating columns, and allows gentler elution conditions with added flexibility in the elution buffer composition.
Sample data provided below demonstrates the improvement in purity when using tag-removal protease elution methods (Figure 3 & 4). The His-tagged protein in this example was eluted either using imidazole, or by on-column cleavage using HRV-3C protease, biotin-tagged. As seen in the SDS-PAGE analysis, on-column elution yields a higher purity final protein with less impurities visible on the gel.
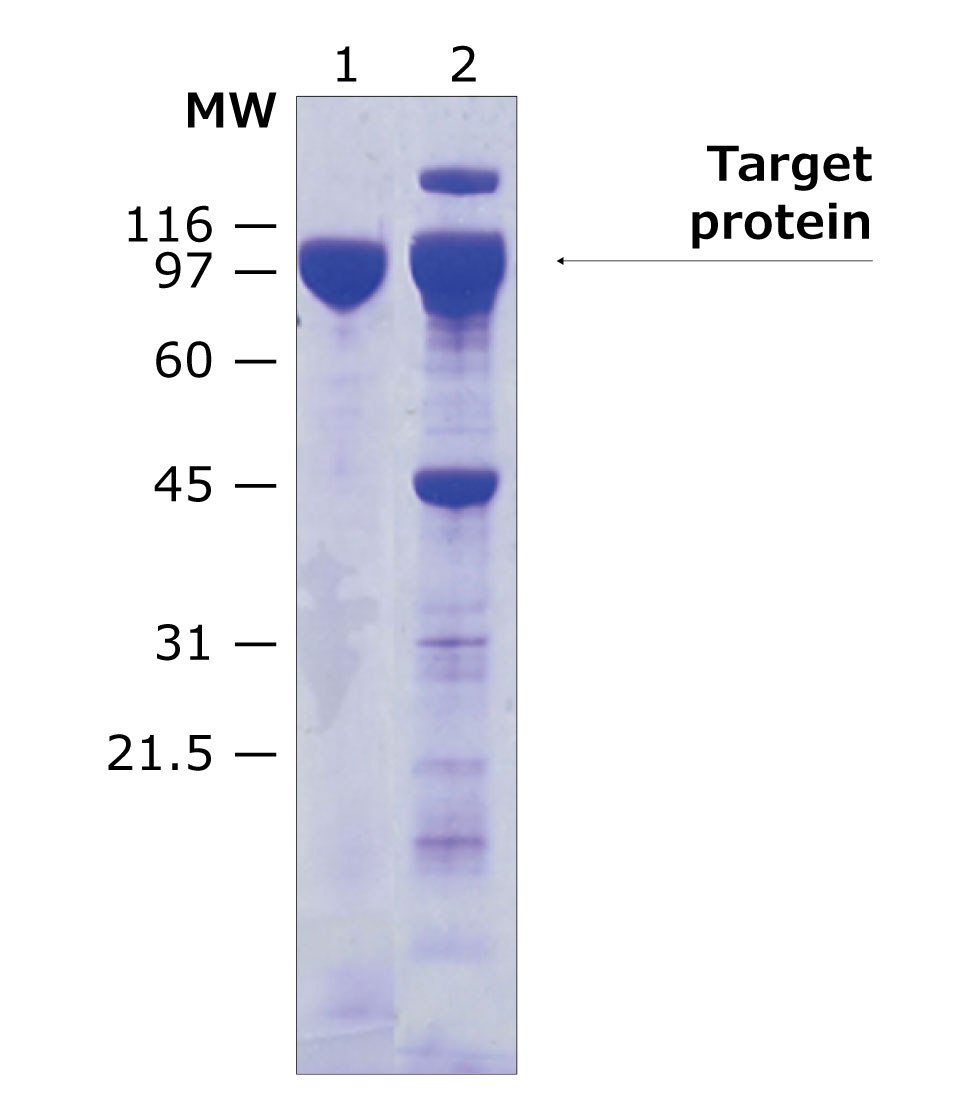
Figure 3.SDS-PAGE analysis of target protein purification using on-column protein cleavage. Lane 1: Purified target protein eluted by on-column cleavage using biotin-tagged HRV-3C protease (Cat. No. SAE0110), Lane 2: purified target protein using imidazole (Cat. No. I202).

Figure 4.The biotinylation degree of thrombin protease (Cat. No. SAE0147), biotin-tagged as analyzed by gel shift assay. Lane 1: Gel shift with streptavidin, Lane 2: No streptavidin.
For additional biotin-tagged protein purification guides, please explore our Purification or Removal of Biotin and Biotinylated Substances and our Purification or Removal of Biotin and Biotinylated Biomolecules with Magnetic Beads resource pages.
Efficiently removing biotinylated proteins
Importantly, it is critical that the degree of biotinylation is high enough to effectively remove the biotin-protease. Any non-biotinylated protease will not bind to the streptavidin and will thus not be removed from the final pure protein preparation. Each batch of our catalog products of biotin-tagged proteases is tested for the degree of biotinylation to verify that it is ≥90%. This quality check ensures that once streptavidin is added that the protease can be efficiently removed.
Related Products
To continue reading please sign in or create an account.
Don't Have An Account?