YeastBuster™ Protein Extraction Reagent for Fast, Efficient Extraction of Proteins from Yeast
The yeast widely studied as a model eukaryotic system. The availability of the complete genome sequence, Saccharomyces cerevisiae is expression libraries, and deletion strains provide necessary tools for genetic screens and selections.1 Yeasts are capable of critical posttranslational modifications of recombinant proteins such as glycosylation, acetylation, phosphorylation, lipidation, cofactor uptake, and protein processing to remove leader and signal sequences.2 S. cerevisiae, Pichia pastoris, Hansulae polymorpha, and Kluvyeromyces lactis have been frequently used to take advantage of these modifications and express functional proteins.3 Polyhistidine (His•Tag® sequence) fusion technologies have been employed in yeast to facilitate protein purification.4,5 However, yeast cell lysis is problematic due to the thick recalcitrant cell wall that can compose approximately 25% of the cells’ dry weight. The yeast cell wall is comprised primarily of polysaccharides and glycoproteins with a high proportion of carbohydrate. Typical components are β-1,3- and β-1,6-D-glucans, β-1,3- and β-1,4-D-glucans, cellulose, mannoproteins, and chitin. This combination of polymers and proteins is interconnected by a combination of covalent, disulfide, hydrogen, and hydrophobic bonds.6 There are numerous methods for disruption of yeast cells, ranging from enzymatic digestion with β-1,3-glucanases to mechanical disruption with various hardware, pressure disruption, freeze fracture and glass bead abrasives combined with mechanical and enzymatic protocols. S. cerevisiae cell lysis has also been controlled through manipulation of genes involved in cell wall biogenesis.7 All of these methods will result in some disruption, but depending on the cell type and culture age, the degree of effective breakage is unpredictable and usually determined by trial and error. Additionally, vigorous mechanical treatment often results in heat and oxidative degradation of proteins.
Recently, extraction and recovery of proteins from both prokaryotes and eukaryotes have been simplified through the development of specialized reagents that eliminate the need for harsh mechanical disruption. Proteins can be extracted from E. coli cell pellets with BugBuster® Protein Extraction Reagent,8,9 or they can be extracted directly from the total culture without cell harvest, mechanical disruption, or extract clarification with PopCulture™ Protein Extraction Reagent.10,11 CytoBuster™ and Insect PopCulture Reagents have been developed for extraction of proteins from cultured mammalian and insect cells. Here we introduce YeastBuster™ Protein Extraction Reagent, a specially formulated mixture of detergent, protein stabilization buffer, and tris(hydroxypropyl)phosphine (THP) reducing agent. This powerful combination eliminates the inconsistencies associated with tedious mechanical and enzymatic lysis and provides a fast, efficient, and gentle extraction method to obtain soluble active proteins from yeast cells.
Comparison with other extraction methods
YeastBuster™ Reagent was compared to two other methods of protein extraction: 1) another commercial yeast protein extraction reagent, and 2) the traditional glass bead method. Log-phase S. cerevisiae cells transformed with an expression vector were harvested by centrifugation and extracted. The extracts generated by the three methods were analyzed by SDS-PAGE, Non-Interfering Protein Assay™ Kit, and enzymatic assays to visualize and quantify soluble and enzymatically active protein.
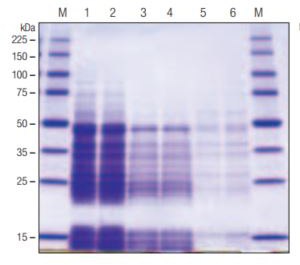
A. SDS PAGE
B. Protein and reporter assays
Figure 1. Performance comparison of YeastBuster™ Protein Extraction Reagent, another commercial reagent, and the glass bead method.
Panel A. SDS-PAGE analysis (4–20% gradient gel) and Coomassie blue staining of extracted proteins. S. cerevisiae cells containing a recombinant plasmid expressing a 35.6 kDa GST•Tag™/His•Tag® fusion protein were grown at 30°C, induced for expression, and harvested at OD600 of 1.2. Cells were collected by centrifugation at 3,000 × g and resuspended in ice cold sterile water. Equal volumes of cells were dispensed into microcentrifuge tubes, and pelleted at 3,000 × g. Cell pellets (~65 mg wet weight) were resuspended in 330 µL of the respective extraction reagents supplemented with 0.5 mM AEBSF and 15 µg/mL benzamidine. The YeastBuster™ Reagent also included 0.01 volume 100X THP Solution as directed in the protocol. After initial resuspension of pellets by pipetting, YeastBuster™ reagent and competitor reagent samples were agitated gently at room temperature for 20 min. Glass bead extraction was accomplished by resuspending the 65-mg pellet in lysis buffer containing 50 mM Tris-HCl, 250 mM LiCl, 100 mM (NH4)2SO4, 1.0 mM DTT, and 2% glycerol, adding approximately 50 µL acid-washed glass beads (100–150 µm diameter), and vortexing the sample on high for 4 min with intermittent chilling on ice. All samples were centrifuged at 16,000 × g for 5 min before SDS-PAGE analysis.
Panel B. Analysis of total protein and reporter activities. Total protein extracted by the three methods was determined using Calbiochem’s Non-Interfering Protein Assay™ Kit. Samples were 10 µL YeastBuster™ reagent extract, 10 µL competitor reagent extract, and 50 µL glass bead extract. Assays were performed in duplicate according to kit protocol. GST activity was determined using the GST•Tag Assay Kit. Assays were performed in duplicate using 5 µL extracted protein from the samples prepared as described for Panel A. The negative control was a S. cerevisiae host carrying the same expression vector, but encoding the lacZ gene instead of GST. Data reflect the average of duplicate assays. β-gal activity was determined using the host expressing lacZ. Cells were grown and processed as described for Panel A. Samples (5 µL) of the extracts were assayed in duplicate using the BetaRed™ β-Gal Assay Kit. The negative control was the extract from the GST-expressing host. Data reflect the average of duplicate assays.
The SDS-PAGE analysis (Figure 1A) shows the superior protein extraction efficiency of YeastBuster™ reagent over the same molecular mass range. The protein assay data (Figure 1B) show that YeastBuster™ reagent extracted nearly twice as much protein as the competitor yeast protein extraction reagent, and nearly ten-fold more than the glass bead method. Reporter assays were also performed to measure activities of the recombinant glutathione-S-transferase (GST) and β-galactosidase (β-gal) in the extracts. The activity data for GST show that YeastBuster™ reagent extracts contained nearly three-fold more activity than competitor reagent extracts and approximately ten-fold more activity than extract prepared with glass beads. YeastBuster™ reagent prepared extract of a host expressing recombinant β-gal had greater than 40-fold more active enzyme than extract prepared with the competitor reagent. Glass bead-prepared extract from the same cells contained approximately two-fold greater β-gal activity than YeastBuster™ reagent.
Because the components in YeastBuster™ reagent or the competitor reagent do not inhibit β-gal activity (data not shown), it is possible that a very large protein such as the β-gal tetramer (ca. 465,000 Da) is not easily extracted from the yeast cell by chemical methods due to physical entrapment or adsorptive interaction with insoluble cellular components. Glass beads may facilitate extraction of β-gal by physical shearing of cellular debris and therefore minimize entrapment. Smaller proteins such as GST would be less likely to suffer from these interactions, and as a result are easier to extract chemically. The advantages of speed, convenience and biocompatibility that YeastBuster™ reagent offers over glass beads and the competitor reagent make it the most attractive overall choice for extraction of proteins. The YeastBuster™ reagent method also enables simple processing of multiple samples, small samples, and other applications that would be extremely tedious or impractical using glass beads.
The activity data for GST show that YeastBuster™ reagent extracts contained nearly three-fold more activity than competitor reagent extracts and approximately tenfold more activity than extract prepared with glass beads. YeastBuster™ reagent prepared extract of a host expressing recombinant β-gal had greater than 40-fold more active enzyme than extract prepared with the competitor reagent.
Compatibility of YeastBuster™ reagent extracts with GST•Bind™ and Ni-NTA His•Bind® Resins
Compatibility of YeastBuster™ reagent extracted proteins for purification by Ni-NTA His•Bind® immobilized metal affinity chromatography (IMAC) and GST•Bind™ affinity methods is shown in Figure 2. A soluble extract was prepared from log-phase S. cerevisiae expressing the GST•Tag™/ His•Tag® fusion protein. Samples of the extract were subjected to GST•Bind™ and Ni-NTA His•Bind® chromatography. Anaysis of purified samples by SDS-PAGE clearly demonstrates that YeastBuster™ is compatible with high-yield affinity purification using both of these methods. Although high concentrations of 2-mercaptoethanol or dithiothreitol are typically used in yeast lysis buffers to reduce disulfide bonds in the cell wall, these are not compatible with IMAC due to reduction and leaching of the complexed Ni2+. The reducing agent THP (provided as a separate 100X stock solution with YeastBuster™ reagent) is effective at concentrations low enough to be compatible with the Ni-NTA His•Bind® chromatography while effectively enhancing yeast cell lysis, even in stationary phase cultures where cell wall thickness and bud scars make lysis increasingly difficult. YeastBuster™ reagent without THP addition will lyse S. cerevisiae harvested in early log phase; however, total protein extraction efficiency will be decreased.

Figure 2. SDS-PAGE analysis of GST•Bind™ and Ni-NTA His•Bind® purified samples.
S. cerevisiae cells containing a recombinant plasmid expressing a 30.5 kDa GST•Tag™/His•Tag® fusion protein were grown at 30 °C, induced for expression, and harvested at OD600 of 1.2. The culture was centrifuged at 3000 × g for 10 min to collect the cells. The pellet (2 g wet weight) was resuspended by briefly pipetting and vortexing in 10 mL YeastBuster™ Reagent containing 0.01 vol 100X THP Solution, 125 U Benzonase® Nuclease, 0.5 mM AEBSF and 15 µg/mL benzamidine, then gently agitated for 20 min. The sample was centrifuged at 16,000 × g for 5 min and 4.5 mL aliquots of the supernatant were applied to 1 mL gravity flow columns of equilibrated GST•Bind™ and Ni-NTA His•Bind® Resins. The GST•Bind™ column was washed with 10 mL 1X GST•Bind™ Wash Buffer and eluted with 3 × 1 mL 1X GST•Bind™ Elute Buffer. The Ni-NTA His•Bind® column was washed with 5 mL 1X Ni-NTA Bind Buffer, 15 mL 1X Ni-NTA Wash Buffer, and eluted in 3 × 1 mL 1X Ni-NTA Elute Buffer. The protein content of the eluates was determined by BCA and Coomassie assays and duplicate samples were analyzed by SDS-PAGE (4–20% gradient gel) and Coomassie blue staining. Lanes are indicated.
Summary
Through the application of YeastBuster™ Protein Extraction Reagent, it is possible to rapidly and efficiently extract and recover proteins from yeast cells without exposing them to the harsh conditions associated with abrasive grinding, ultrasonication and pressure disruption. Release of soluble active proteins is rapidly achieved with YeastBuster™ Reagent, and inconsistencies resulting from tedious mechanical and enzymatic lysis are eliminated. In addition to greater total protein yields in crude extracts and recovery of enzymatically active protein, recombinant fusion proteins can be affinity purified using GST•Bind™ and Ni-NTA His•Bind® resins.
Materials
如要继续阅读,请登录或创建帐户。
暂无帐户?