Preactivated Magnetic Beads for Affinity Chromatography
NHS Mag Sepharose, Sera-Mag Carboxylate-Modified, Sera-Mag SpeedBeads Carboxylate-Modified
Preactivated magnetic beads are designed for covalent coupling of antibodies, aptamers, and proteins. After coupling is performed, proteins of interest can be affinity captured and enriched using immunoprecipitation (example in Figure 5.7). The preactivated magnetic beads include NHS Mag Sepharose, Sera-Mag Carboxylate-Modified, and Sera-Mag SpeedBeads Carboxylate-Modified Magnetic Particles.

Figure 5.7.Principle for coupling of biomolecules.
NHS Mag Sepharose has an N-hydroxysuccinimide ligand to which molecules with primary amino groups bind covalently. This enables enrichment of target protein for further downstream analyses such as LC-MS and electrophoresis techniques.
Sera-Mag and Sera-Mag SpeedBeads Carboxylate-Modified Magnetic Particles have carboxylic groups on the surface that permit easy covalent coupling using simple carbodiimide chemistry.
Proteins bind to carboxylate-modified particles by adsorption. Adsorption is mediated by hydrophobic and ionic interactions between the protein and the surface of the particles.
In addition to being adsorbed, proteins can be covalently attached to the surface of carboxylate-modified particles. Carboxyl groups on the particles, activated by the water-soluble carbodiimide 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC), react with free amino groups of the adsorbed protein to form amide bonds.
Bead characteristics
Characteristics of Mag Sepharose, Sera-Mag, and Sera-Mag SpeedBeads preactivated magnetic beads are shown in Table 5.3.
Purification example
Enrichment of plasminogen from human plasma
Human plasma contains a vast number of proteins and can be difficult to work with due to the great range of protein concentrations. In this experiment, plasminogen was enriched from human plasma by immunoprecipitation. Monoclonal antiplasminogen mouse IgG1 was covalently coupled to NHS Mag Sepharose for capture of plasminogen. The eluted fractions were analyzed by SDS-PAGE (Figure 5.8) and showed efficient enrichment of the target protein. Enrichment and identification of plasminogen were confirmed by LC-MS/MS analysis.

Figure 5.8.SDS-PAGE results of the enrichment of plasminogen from human plasma using NHS Mag Sepharose magnetic beads. The gel was post-stained with Deep Purple Total Protein Stain and scanned.
Performing a separation
NHS Mag Sepharose
The optimal parameters for protein enrichment are dependent on the specific combination of biomolecules used. Optimization may be required for each specific combination to obtain the best result. Examples of parameters which may require optimization are:
- Amount of beads
- Amount of antibodies
- Amount of protein (antigen) to be enriched
- Incubation times
- Choice of buffers
- Number of washes
Antibody solution preparation
Prepare the antibody solution by dilution in coupling buffer and keep it on ice.
Coupling and purification of target protein
Use the magnetic rack with the magnet in place to attract the beads before each liquid removal step.
- Prepare the Mag Sepharose beads
- Mix the medium slurry thoroughly by vortexing. Dispense 25 µL of medium slurry into an Eppendorf tube.
- Place the Eppendorf tube in the magnetic rack to attract the beads.
- Remove the storage solution.
- Equilibration
- Add 500 µl ice cold equilibration buffer and resuspend the medium.
- Remove the liquid.
- Binding of antibody
- Immediately after equilibration, add the antibody solution (at least 50 µl).
- Resuspend the medium and incubate at least 15 min with slow end-over-end mixing or by using a benchtop shaker.
- Remove the liquid.
- Blocking of residual active groups
- Add 500 µl blocking buffer A and remove the liquid.
- Add 500 µl blocking buffer B and remove the liquid.
- Add 500 µl blocking buffer A.
- Incubate for 15 min with slow end-over-end mixing.
- Remove the liquid.
- Add 500 µl blocking buffer B and remove the liquid.
- Add 500 µl blocking buffer A and remove the liquid.
- Add 500 µl blocking buffer B and remove the liquid.
- Equilibration for binding
- Add 500 µl binding buffer and resuspend the medium.
- Remove the liquid.
- Binding of target protein
- Add sample, diluted in, for example, binding buffer.
- Resuspend the medium and incubate for 10 to 60 min with slow end-over-end mixing.
- Remove the liquid.
- Wash (perform this step three times)
- Add 500 µl wash buffer.
- Remove the liquid.
- Elution (perform this step twice)
- Add 50 µl elution buffer.
- Resuspend the medium and incubate for at least 2 min.
- Remove and collect the elution fraction. The collected elution fraction contains the main part of the protein. If needed, repeat the elution.
Do not use an amine-containing buffer (e.g. Tris or glycine) for the antibody/protein that is to be coupled since the amines in the buffer will compete for the coupling sites.
Remove potential amines before coupling with antibody solution, for example by dialysis or buffer exchange with a desalting column.
Sera-Mag and Sera-Mag SpeedBeads Carboxylate-Modified
Sera-Mag and Sera-Mag SpeedBeads Carboxylate-Modified Magnetic Particles feature carboxylic groups on the surface that permit easy covalent coupling using simple carbodiimide chemistry (Figure 5.9).
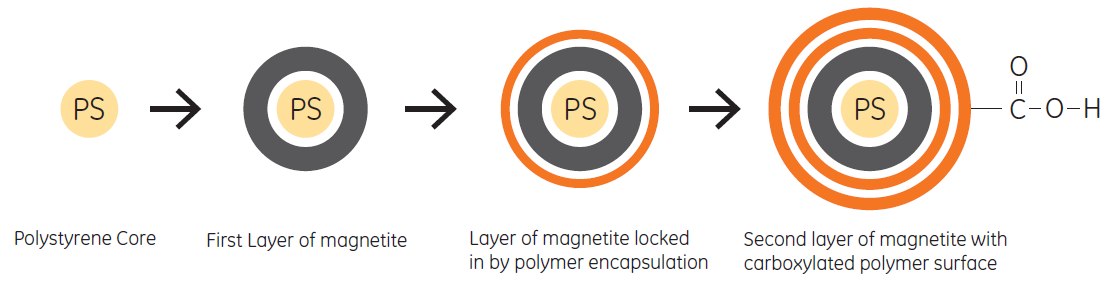
Figure 5.9.Sera-Mag SpeedBead Carboxylate-Modified Magnetic Particles feature carboxylic groups on the surface that permit easy covalent coupling using simple carbodiimide chemistry.
Two procedures exist for coupling of proteins and there is also a method for coupling of oligonucleotides. The one-step covalent coupling procedure with EDAC is recommended for covalent coupling of most proteins. However, if EDAC is found to damage the protein of interest, the two-step procedure, which includes a preaactivation (active ester) step prior to introducing the protein, can be used for covalent coupling.
EDAC is very sensitive to moisture and undergoes rapid hydrolysis in aqueous solutions. Therefore, EDAC should be stored in a desiccator at -20 °C and brought to room temperature just before weighing.
Sample preparation
The protein used to coat the particles should be completely dissolved and not too concentrated. A concentration of 1 to 10 mg/mL in water is recommended for most proteins.
Optimizing the amount of EDAC and protein
- Determine the optimal molar ratio of EDAC:COOH.
- For one-step coupling: Perform an EDAC titration while holding the amount of protein constant. We recommend EDAC:COOH ratios of 0, 0.5, 1, 2.5, 5 and 10:1 for optimization.
For two-step coupling: Optimize the EDAC:COOH ratio, starting with a recommended 2.5:1 ratio. A molar ratio of 20:1 NHS:COOH is recommended for all reactions.
- Determine the amount of protein to add. Perform a protein titration, holding the determined EDAC concentration fixed.
The optimal amount of protein to use depends on several factors:- Surface area available: surface area/mg of particles increases linearly with decreasing particle diameter.
- Colloidal stability: proteins can have stabilizing or destabilizing effects on the particles.
- Immunoreactivity: the optimal amount of bound sensitizing protein must ultimately be determined by functional assay. Performing a protein titration or binding isotherm is a good first experiment. It is recommended to start with protein concentrations of 0, 25, 50, 75, 100, and 150 or 200 µg/mg of particle.
Calculation of required amount of EDAC
Use the following information to calculate the amount of EDAC required. Optimizing the amount of EDAC and protein above for details. Reactions of 1 mL are recommended for optimization. The magnetic bead acid content, provided in mEq/g, is equivalent to µmol/mg. Note! 1 mL of 1% medium slurry contains 10 mg of magnetic beads.
The µmol EDAC required = (acid content, in µmol/mg) × 10 mg of magnetic beads × desired ratio. Use this value in the following equation to determine how much of the EDAC stock to add:
| µmol EDAC required | = mL of EDAC stock for a 1 mL reaction |
| 52 µmol/mL |
One-step coupling procedure for proteins
Use the magnetic rack with the magnet in place to attract the beads before each liquid removal step.
- Prepare Sera-Mag beads
- Mix the 10% medium slurry throughly by vortexing. Dispense 100 µL of the homogenous medium slurry into an Eppendorf tube.
- Add 50 µL of 10× MES buffer (to 25 mM final concentration) and water to bring reaction up to 1.0 mL final volume.
- Add protein stock solution (Optimizing the amount of EDAC and protein for details)
Resuspend the medium and incubate for approximately 15 min by end-over-end mixing or a benchtop device.
- Activation and coupling
- Prepare the EDAC solution immediately before use.
- Mix the calculated volume of EDAC solution rapidly into the reaction by pipetting up and down repeatedly.
- Incubate for 1 h by end-over-end mixing. The beads may flocculate during this time, but this is not unusual or harmful.
- Perform centrifugation (10 to 30 min in a standard microcentrifuge)
Decant the supernatant.
- Wash (perform this step twice)
- Add 50 µL of 10× MES buffer (to 25 mM final concentration) and water to bring reaction up to 1.0 mL, or use a higher pH buffer of choice. Use ultrasonication for resuspension.
- Perform centrifugation as in previous step.
Decant the supernatant.
- Resuspension
- Add buffer that does not contain blocking proteins (use MES buffer or a higher pH buffer of choice)
Resuspend the coupled magnetic beads to the desired medium slurry concentration.For example, if the target of solid concentration is 1.0%, add 0.97 mL of the buffer, which accounts for a small amount of liquid that will remain after pellet formation.
The amount of protein bound on the magnetic beads is determined by a BCA protein assay. For long-term colloidal stability, a stabilizing storage buffer will be needed. After performing the protein analysis, coated beads can be centrifuged and resuspended in a variety of storage buffers, and the colloidal stability and reactivity can be optimized.
Covalently bound protein will not elute when subjected to detergent washes or buffer changes. Thus, covalently coupled reagents are compatible with a wide variety of buffer additives.
Active ester two-step coupling procedure for proteins
Use the magnetic rack with the magnet in place to attract the beads before each liquid removal step.
- Prepare Sera-Mag beads
- Mix the 10% medium slurry throughly by vortexing. Dispense 100 µL of the homogenous medium slurry into an Eppendorf tube.
- Add 100 µL of 10× MES buffer (to 50 mM final concentration) and water to bring reaction up to 1.0 mL final volume.
Add 230 µL NHS solution: 100 mM final concentration and resuspend the medium.
- Activation
- Prepare the EDAC solution immediately before use.
- Mix the calculated volume of EDAC solution rapidly into the reaction by pipetting up and down repeatedly.
- Incubate for 30 min by end-over-end mixing. The beads may flocculate during this time, but this is not unusual or harmful.
- Perform centrifugation (10 to 30 min in a standard microcentrifuge).
Decant the supernatant.
- Wash (perform this step twice)
- Add 1 mL of 50 mM MES buffer, pH 6.1. Use ultrasonication for resuspension.
- Perform centrifugation as in previous step.
Decant the supernatant.
- Coupling
- Add the protein stock solution (Optimizing the amount of EDAC and protein for details)
- Resuspend the medium and incubate for 1 h by end-over-end mixing.
- Perform centrifugation as in previous step.
Decant the supernatant.
- Wash (perform this step twice)
- Add 100 µL of 500 mM MES buffer and fill up to 1 mL with water. Use ultrasonication to resuspend pellet.
- Perform centrifugation as in previous step.
Decant the supernatant.
- Resuspension
- Add buffer that does not contain blocking proteins (use MES buffer or a higher pH buffer of choice)
Resuspend the coupled magnetic beads to the desired medium slurry concentration. For example, if the target concentration for solids is 1.0%, add 0.97 mL of the buffer, which accounts for a small amount of liquid that will remain after pellet formation.
The amount of protein bound on the magnetic beads is determined by a BCA protein assay. For long-term colloidal stability, a stabilizing storage buffer will be needed. After performing the protein analysis, coated beads can be centrifuged and resuspended in a variety of storage buffers, and the colloidal stability and reactivity can be optimized.
Note that covalently bound protein will not elute when subjected to detergent washes or buffer changes. Thus, covalently coupled reagents are compatible with a wide variety of buffer additives.
Use the magnetic rack with the magnet in place to attract the beads before each liquid removal step.
- Prepare the Sera-Mag beads
- Mix the 10% medium slurry thoroughly by vortexing. Dispense 200 µL of the homogenous medium slurry into an Eppendorf tube.
- Add 100 µL of 10× MES buffer (to 50 mM final concentration)
- Add amine-modified oligonucleotide modified in water
Add DNase/RNase-free water to 1 mL final volume
- Activation and coupling
- Prepare the EDAC solution immediately before use.
- Add 100 µL EDAC solution rapidly into the reaction by pipetting up and down repeatedly.
- Incubate for overnight by end-over-end mixing at 37 °C. The beads might flocculate during this time, but this is not unusual or harmful.
Remove the liquid.
- Wash 1 (perform this step twice)
- Add 1 mL of DNase/RNase-free water and resuspend the medium.
Remove the liquid.
- Wash 2 (perform this step twice)
- Add 1 mL of 100 mM imidazole (pH 6.0).
- Resuspend the medium and incubate for 5 min at 37 °C by end-over-end mixing.
Remove the liquid.
- Wash 3 (perform this step twice)
- Add 1 mL of 100 mM sodium bicarbonate.
- Resuspend the medium and incubate for 5 min at 37 °C by end-over-end mixing.
Remove the liquid.
- Wash 4 (perform this step twice)
- Add 1 mL of 100 mM sodium bicarbonate.
- Resuspend the medium and incubate for 30 min at 65 °C by end-over-end mixing.
Remove the liquid.
- Resuspension
- Add DNase/RNase-free water or buffer of choice for storage.
Materials
如要继续阅读,请登录或创建帐户。
暂无帐户?