Induced Pluripotent Stem Cell Reprogramming Protocols
Introduction
Methods
Reprogramming of Human Fibroblasts using Non-Integrating Self-Replicating RNA Vectors
Reprogramming of Peripheral Blood Mononuclear Cells (PBMCs) using STEMCCA Lentiviral Vectors
Characterization and Picking of Reprogrammed iPSC Colonies
Frequently Asked Questions
Materials
Introduction
A ready source of induced pluripotent stem cells (iPSCs) is critical to the effective study of differentiation pathways and the investigation of the therapeutic potential of iPS cells. Since the discovery that human iPSCs could be generated by inducing expression of the four reprogramming factors (OCT-4, SOX-2, KLF-4 and c-MYC)1 many different reprogramming technologies have emerged to generate iPSCs, each possessing their own advantages and disadvantages2.
First-generation technologies, based on retroviral and lentiviral systems, allowed for highly efficient reprogramming events but lacked the necessary control over host genome integrations. Cre-excisable lentiviral systems offered a solution to genome integration but required lengthy subcloning procedures and screening to ensure excision of the reprogramming factors.
Second-generation technologies used non-integrating episomal DNA plasmids, which were transgene-free but lacked the high reprogramming efficiencies of earlier retroviral and lentiviral techniques. Third-generation technologies used negative sense, non-integrating RNA viruses, termed Sendai Viruses (SeV), which originated from highly transmissible respiratory tract infections in mice, hamsters, guinea pigs, rats, and pigs. These RNA viruses produced integration-free iPSCs, produced high reprogramming efficiencies and were easy to use, but residual Sendai virus was difficult to clear from cells, resulting in the requirement for multiple rounds of clonal expansion and analysis.
Next generation reprogramming system that use synthetic self-replicating RNA engineered to mimic cellular RNA have been used to generate human iPS cells3. The single RNA strand contains the four reprogramming factors and enables extremely efficient reprogramming using a single transfection step without any viral intermediates or host genome integration. Once iPSCs are generated, the RNA can easily be selectively eliminated by removing the interferon-gamma (IFNg) inhibitor, B18R, from the cell culture medium.
Browse all Pluripotent Stem Cell Products
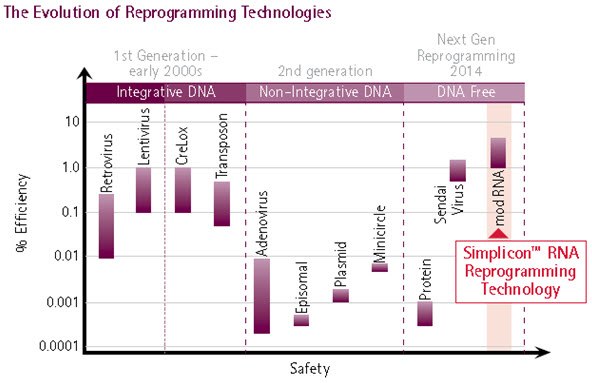
Figure 1. Evolution of Reprogramming Method.The evolution of reprogramming technologies has culminated in the development of synthetic RNA-mediated reprogramming (extreme right), representing the safest and most efficient method for iPS cell generation: Simplicon™ RNA Reprogramming Technology combines the efficiency of retroviral and lentiviral reprogramming technologies with the safety of non-viral based reprogramming methods.
Figure 2. Simplicon RNA Human HFF Reprogramming Timeline.Using a single transfection of self-replicating RNA can produce high numbers of integration-free iPSCs following a 3-4 week protocol.
Before Starting Experiment:
- Determine the optimal plating density of target human fibroblast cells (SCC058). The optimal plating density is defined as the number of cells that should be plated at Day 0 in order to have the cells reach 60- 80% confluency the next day (Day 1). Plate out a range of cell numbers from 1 x 105 to 1 x 106 cells per well of a 6-well plate in fibroblast culture media (SCM044)
- Determine the optimal starting puromycin (P8833) concentration. The optimal starting puromycin concentration is defined as half the concentration required to achieve 50% cell death by Day 4-5. Puromycin sensitivity may vary based on the target cell line and must be determined empirically before starting.
Reprogramming Protocol
Day 0: Plate target Cells
- Plate target cells at the optimal plating density (determined from Step 1) in the same culture medium that was used to maintain target cells in the proliferative state. Volume should be 3 mL per well of a 6-well plate. Set aside an untransfected control well to observe the puromycin cell death.
- Thaw the following Simplicon RNA Reprogramming Kit (SCR550) components on ice and aliquot into sterile nuclease-free Eppendorf tubes. Store at -80 °C until ready to use.
• VEE-OKS-iG RNA: Aliquot 3 µL into sterile nuclease-free eppendorf tubes. Store aliquots at -80 °C.
• B18R RNA: Aliquot 3 µL into sterile nuclease-free eppendorf tubes. Store aliquots at - 80 °C.
• B18R protein: Aliquot 3 µL into sterile nuclease-free eppendorf tubes. Store aliquots at - 80 °C.
Day 1: Pretreat cells with B18R protein Transfect with Simplicon™ VEE-OKS-iG and B18R RNA
- On the day of transfection, cells should be 60-80% confluent. Pre-treat target cells for two hour with the 200ng/mL B18R protein (GF156) in 1mL/well DMEM (SLM-120-B) to help suppress the cellular interferon response before transfection with the Simplicon™ RNAs.
- Prepare the RNA/Transfection complex by combining 0.5ul VEE-OKS-iG and B18R RNAs and 4.0ul RiboJuice™ mRNA Transfection Reagent (TR-1013) in 250ul Opti-MEM media. Add the RNA/Transfection reagent complexes drop wise into one well of the 6-well plate containing cells, mix by rocking and incubate the plate in a 37˚C, 5% CO2 incubator for 4 h.
- After 4 hours replenish media with DMEM (SLM-120-B), 10% FBS (ES-009-B), 1X Glutamine (A8185) and 200ng/mL B18R protein (GF156) and incubate in a 37˚C, 5% CO2 incubator.
Day 2-Day 11: Apply Optimal Starting Puromycin Concentration
- Aspirate the medium. Replace with 2 mL of FBS media containing 200 ng/mL B18R protein (see step 7 for formulation). Add in the optimal starting puromycin (P8833) concentration that was determined from step 2. Gently rock the plate from side to side to thoroughly mix the puromycin to the medium.
- Replace the medium daily with 2 mL Stage 1 medium containing 200 ng/mL B18R protein and fresh puromycin.
Note: Monitor daily to assess the cell’s response to the puromycin.
- By day 4 – 5, variable levels of cell death may be observed. Adjust the puromycin concentration so that 30-60% of the cells die. Maintain this concentration for up to day 11.
Day 11-18: Replating Reprogrammed Cells and iPSC Expansion
- When puromycin-resistant cells are 70-90% confluent at around day 11, they can be replated onto Matrigel coated plates (for feeder-free culture) or onto inactivated MEF feeder layer (for feeder-based culture). This may occur anytime between days 9 - 18 depending upon the cell’s response to puromycin. Suggested expansion media is MEFCM (SCM103), 10ng/mL bFGF (GF003), 1X Human iPSC Reprogramming Boost Supplement II (SCM094) and 200ng/mL B18R protein (GF156).
Day 18-30: iPSC Colony Picking and Expansion
- Continue to monitor the growth of the human iPSC colonies daily. Look for homogeneous colonies that are compact and have defined borders. When iPSC colonies reach approximately 200 cells or over in size, they are ready to be picked and expanded in feeder-free PluriSTEM Human ES/iPSC Media (SCM130) for further experiments.
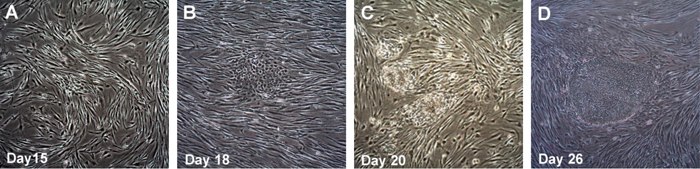
Figure 3. Time course of human iPSC colonies generation using Human Simplicon RNA Reprogramming Kit.The transfected HFFs were replated onto inactive MEFs at Day 10, Colonies start to emerge from Day 15-16 and are more obvious around day 17-20 (A, B). Colonies are ready to be picked at Day 26.
Reprogramming of Peripheral Blood Mononuclear Cells (PBMCs) using STEMCCA Lentiviral Vectors
Day 0: Isolation and Expansion of Peripheral Blood Mononuclear Cells (PBMCs)
- Draw 4 mL of peripheral blood into a BD Vacutainer CPT Cell Preparation Tube with sodium citrate. Invert the tube 8 to 10 times and centrifuge at 1,800 x g for 30 min at room temperature. Ideally, this step should be done within 2 hr of collection.
- Collect the mononuclear cells (MCs) by pipetting the buffy coat (cell layer between gel barrier and plasma) into a sterile 15 mL conical centrifuge tube. Bring total volume to 10 mL with sterile phosphate-buffered saline (D8537) invert several times and centrifuge at 300 x g for 15 min.
- Resuspend the cells in 10 mL of sterile PBS (D8537) and perform cell count. Transfer 1 to 2x106 cells into a sterile 15 mL conical centrifuge tube and centrifuge at 300 x g for 10 min.
- Resuspend the cells in 2 mL of expansion medium (EM) (QBSF-60 Stem Cell Medium containing 50 μg/mL Ascorbic Acid (A4403) , 50 ng/mL SCF (S7901), 10 ng/mL IL-3 (I1646), 2 U/mL EPO (H5166), 40 ng/mL IGF-1 (I3769), 1 μM Dexamethasone (D4902) and 1% Pen/Strep (P4333) and transfer to one well of a 12-well plate. Incubate the cells in a 37 °C, 5% CO2 incubator.
- Centrifuge remaining cells at 300 xg for 10 min and freeze ~2x106 cells/vial in FBS containing 10% DMSO (D2650).
- To start the protocol using frozen PBMCs, thaw 1 vial of cells into 10 mL of QBSF medium and centrifuge at 300 x g for 10 min. Resuspend the cells in 2 mL of EM and transfer to one well of a 12-well plate. Incubate the cells in a 37 °C, 5% CO2 incubator.
Day 3 and 6
- Transfer the cells to a sterile 15 mL conical tube and wash the well once with 1 mL of QBSF-60 Stem Cell Medium to collect adherent cells.
- Centrifuge the cells at 300 x g for 10 min.
- Resuspend the cells in 2 mL of EM and transfer to one well of a 12-well plate. Incubate the cells in a 37 °C, 5% CO2 incubator.
Day 9: Transduction of PBMCs with STEMCCA Lentivirus
- Transfer the cells to a sterile 15 mL conical tube and wash the well once with 1 mL of QBSF-60 Stem Cell Medium to collect adherent cells.
- Centrifuge the cells at 300 x g for 10 min.
- Resuspend the cells in 1 mL of fresh EM containing 5 μg/mL of polybrene (TR-1003) and STEMCCA lentivirus (SCR544) (MOI=1 to 10) and transfer to one well of a 12-well plate.
- Spin the plate at 2,250 rpm at 25 °C for 90 min.
- After spin, add an additional 1 mL of fresh EM containing 5 μg/mL of polybrene for a total of 2 mL of medium and incubate the plate in a 37 °C, 5% CO2 incubator.
Day 10
- Transfer the cells to a sterile 15 mL conical tube and wash the well once with 1 mL of QBSF-60 Stem Cell Medium to collect adherent cells.
- Centrifuge the cells at 300 x g for 10 min.
Day 11: Plating iPSC onto MEFs
- Coat the wells of a 6-well plate with 0.1% gelatin (ES-006) and plate inactivated mouse embryonic fibroblasts (MEFs) at 2x105 cells/well in MEF medium.
- The following day, transfer the iPSCs to a sterile 15 mL conical tube and wash the well once with 1 mL of QBSF-60 Stem Cell Medium to collect adherent cells.
- Resuspend the cells in 3 mL of MEF medium containing 10 ng/mL bFGF (F0291), 50 μg/mL Ascorbic Acid (A4403) , 50 ng/mL SCF (S7901), 10 ng/mL IL-3 (I1646), 2 U/mL EPO (H5166), 40 ng/mL IGF-1 (I3769), 1 μM Dexamethasone (D4902) and 1% Pen/Strep (P4333).
- Plate 1 mL of cells per well of a 6-well plate containing MEFs. Add 1.5 mL of MEF media with bFGF, Ascorbic Acid, and growth factors for a total of 2.5 mL of media/well.
- Spin the plate at 500 rpm at 25 °C for 30 min. Incubate the plate in a 37 °C, 5% CO2 incubator.
- Feed cells every other day with 2.5 mL of MEF media containing 10 ng/mL bFGF and 50 μg/mL Ascorbic Acid (no growth factors). Aspirate and discard floating cells with each feed. Add additional MEFs as needed.
- Reprogrammed iPSC colonies are ready to be picked and expanded in feeder-free PluriSTEM Human ES/iPSC Media (SCM130) for further experiments at around day 18-21.
Characterization and Picking of Reprogrammed iPSC Colonies
Recent findings indicate that fully reprogrammed human iPS cells possess the following characteristics: (1) they downregulate expression of fibroblast marker, CD13 (2) they upregulate expression of pluripotent markers, SSEA-4 and TRA-1-60, (3) they silence the viral transgenes while (4) reactivating endogenous expression of Nanog and (5) they assume a HoeschstDim phenotype. These characteristics have enabled reliable identification and colony picking of fully reprogrammed human iPS cells from a mixed population of partially reprogrammed cells.
Millipore’s Human iPS Selection Kit (SCR502) is a quick, easy and non-invasive method to monitor the pluripotent state of fully reprogrammed human iPS cells using immunocytochemistry (ICC). The Human iPS Selection Kit allows live cell imaging and identification of fully reprogrammed human iPS cells from a heterogeneous population of reprogramming intermediates and enables the selection of human iPS cells that can be further passaged and expanded for downstream applications.
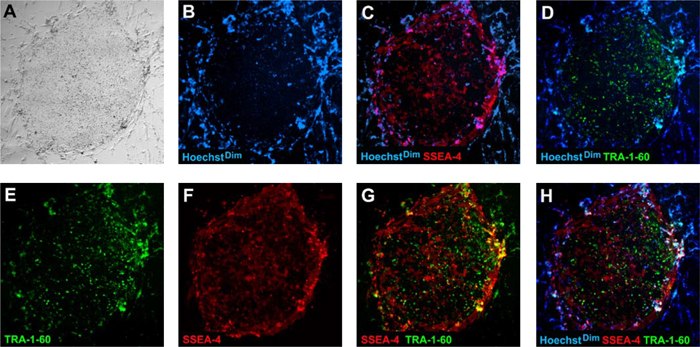
Figure 4.Fully reprogrammed human iPS cells express human pluripotent markers, TRA-1-60 FITC (D, E, G, H, green) and SSEA-4 PE (C, F, G, H, red) while downregulating the fibroblast marker, CD13 PE (data not shown). Cells were stained with cell permeable Hoechst nuclear dye (B, C, D, H, blue). Fully reprogrammed human iPS cells exhibit Hoechst dim phenotype (see colony center in B, C, D, H) while non-iPS and differentiated cells exhibit a Hoechst bright phenotype (see the periphery of the colony in B, C, D, H, which is surrounded by fibroblast cells and are Hoechst bright). Human iPS colonies at passage 5 were used for live staining.
Frequently Asked Questions
- Can Simplicon be used to reprogram PBMCs?
No, the efficiency is too low.
- Do I need to transfect in B18R RNA even though the B18R protein is already included?
We have found empirically that cotransfection of VEE-OKS-iG and B18R mRNA in the presence of the B18R protein increased the reprogramming efficiency compared to using VEE-OKS-iG RNA and B18R protein.
- Is it necessary to add in the small molecules in the Human iPS Reprogramming Boost Supplement II?
No. However, the small molecules dramatically enhances the efficiency and quality of colony formation.
- Can I transfect the RNAs or transduce with Lentivirus more than once? Will it improve my reprogramming efficiency?
Based upon our experience with human foreskin fibroblasts, a single transfection is sufficient. The RNA self-replicates and the puromycin resistance gene will help select for cells that take up the self-replicating RNAs. For other cell types, additional transfections or viral transductions may be helpful but should occur before the replating.
- How long will the RNA self-replicate in the cells?
Based upon PCR data, the RNA is no longer present at p4.
- Do I need to use MEF-CM after replating or can I use another Human ES/iPS Medium?
Other media such as PluriSTEM Media (SCM130) can be used but for best results MEFCM is recommended
- Do I need to replate my transfected cells to MEF feeder layer?
Transfected cells could be replated on MEF feeder layer or on a matrigel coated plate in Stage 2 Medium with B18R protein. B18R protein may be withdrawn when tiny iPS colonies start to emerge.
- Should I use Rock Inhibitor during my replating to increase cell survival?
Rock Inhibitor is not required.
- How can I remove the STEMCCA trasngenes from the host genome?
STEMCCA contains LOXP sites flanking the Yamanaka factors. Once reprogrammed, iPSC cells can be treated with CRE-Recombinase (SCR508) to remove transgenes.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?