HybriScan® Rapid Microbial Test System
The routine control of microbial contamination in food production, from raw materials to finished goods, conventional standard cultivation-based methods are time consuming and often take from 2 to 15 days to get results. Our HybriScan® assays provides fast and accurate molecular detection of spoilage organisms and pathogens, without the need to use expensive PCR methods. HybriScan® Molecular Screening System is suitable for the safety and quality control of beverages, water, and food.
- Rapid Microbiological Method: Sandwich Hybridization
- Sensitivity, Specificity and Flexibility of Sandwich Hybridization
- Advantages Of HybriScan® Technology Over Other Microbial Detection Techniques
- HybriScan® Wastewater Bacterial Count Calculator
- HybriScan®D (Detection) Kits
- HybriScan®I (Identification) Kits
Rapid Microbiological Method: Sandwich Hybridization
The HybriScan® system is based on the detection of microbe-specific rRNA using sandwich hybridization. The method simply needs a centrifuge, a thermal mixer, and an optional microplate reader. The complete analysis can be achieved in 2 hours for the HybriScan®D kit (quantitative kits) and about 1 hour for the HybriScan®I kit (identification kits). Qualitative results can be determined by eye or, for quantitative measurement, used with a microplate reader.
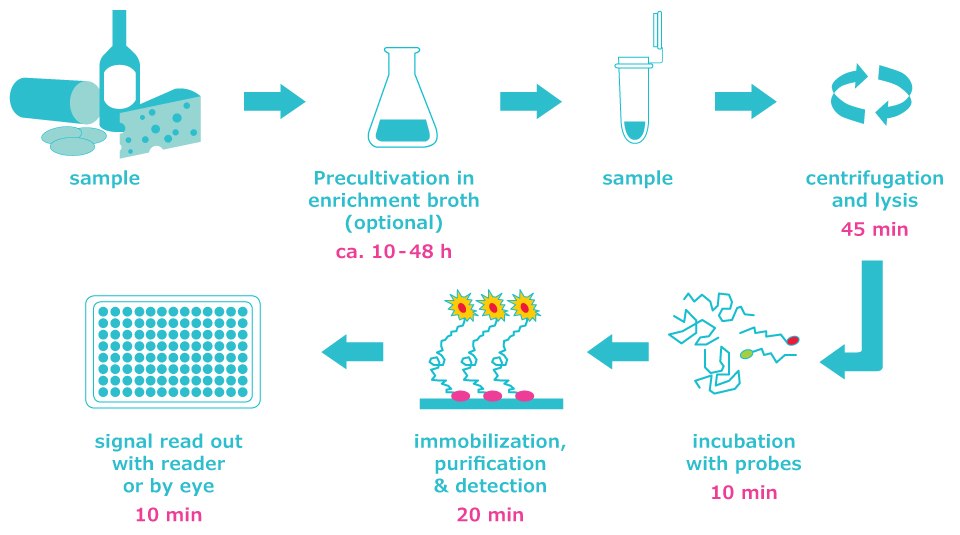
Figure 1.HybriScan® system screening workflow with an analysis time of approx. 2–2.5 hours.
Sensitivity, Specificity and Flexibility of Sandwich Hybridization
Sandwich hybridization is very sensitive and can detect attomoles of the target rRNA molecules in yeast or bacteria.1-3 These types of cells comprise a large number of rRNA-containing ribosomes; a single cell therefore contains several thousand copies of rRNA but only one DNA copy. Sandwich hybridization provides sensitivity in crude biological samples because it is not susceptible to matrix interference. By using specific probes, the HybriScan® system allows both genus or species-specific detection and it is therefore applicable to many analytical fields, including monitoring the microbial content of beer, wine, non-alcoholic beverages, drinking water, a wide variety of foods and wastewater.
Figure 2.HybriScan® technology
- Detects viable microorganisms only—minimizes false-positive results from dead bacteria
- Not sensitive to sample matrix
- Genus- and species-specific detection
- Results in just 2 hours after enrichment
- Broad application range: beer, wine, non-alcoholic beverages, drinking water, food and wastewater
- Can be used to detect non-culturable microbes
HybriScan® Wastewater Bacterial Count Calculator
For our Wastewater Kits - (HybriScan®D Waste Water Microthrix parvicella and HybriScan®D Waste Water Total Bacterial Count), we are providing an excel sheet (below) for calculation of total bacterial count, Microthrix count, and the ratio of Microthrix: Total Bacterial Count.
HybriScan®D (Detection) Kits
HybriScan® I (Identification) Kits
References
To continue reading please sign in or create an account.
Don't Have An Account?