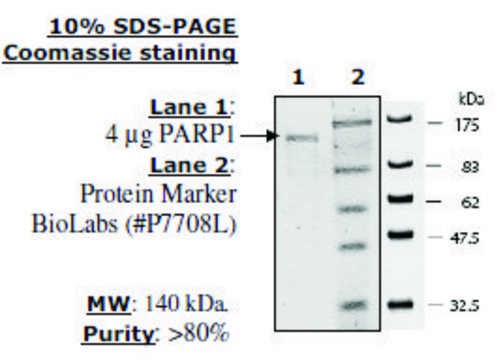
17-10149
PARP1 Enzyme Activity Assay
This PARP1 Enzyme Assay is a flexible & reliable, homogeneous, no-wash assay for quantifying PARP1 & PARP2 activity.
Sign Into View Organizational & Contract Pricing
All Photos(7)
Synonym(s):
poly (ADP-ribose) polymerase-1, ARTD1
UNSPSC Code:
12161503
eCl@ss:
32161000
Recommended Products
manufacturer/tradename
Chemicon®
technique(s)
activity assay: suitable
detection method
fluorometric
shipped in
dry ice
General description
Introduction
Poly(ADP-ribose) (PAR) is a polymer of ADP-ribose present in the nucleus in a free state or as a post-translational modification on glutamate residues on a number of substrates. Synthesis and degradation of PAR are highly regulated, and its presence mediates DNA repair, chromatin structure, cell cycle, and cell death. A family of poly(ADP-ribose) polymerases (PARPs) catalyzes the addition of ADP-ribose moieties onto substrate proteins via cleavage of NAD+ and liberation of nicotinamide. In humans the PARP family is comprised of 17 members, and the members vary significantly in structure and biological function. PARP1 plays a key role in DNA damage repair, particularly base excision repair. It is activated by DNA strand breaks to add PAR to itself and to histones, which results in the recruitment of PAR-binding DNA repair factors, such as XRCC1, to the site of damage. PARP2 overlaps functionally with PARP1 in DNA damage repair, but PARP2 plays a distinct role from PARP1 in development of spermatogonia, adipose tissue, and thymus tissue. Other members of the PARP family are less characterized, and some may function as mono(ADP-ribosyl) transferases rather than polymerases. Consequently, a new nomenclature has been proposed that refers to the common ADP-ribosyltransferase activity of the members.
A number of PARP1 inhibitors have been developed to block repair of chemotherapeutic agent-induced genotoxic damage, and thereby potentiate the cytotoxic effect of chemotherapeutics in cancer patients. Inhibition of PARP1 appears to be particularly efficacious in conjunction with chemotherapeutics on cancer cells harboring mutations in components of homologous recombination DNA repair pathways, primarily BRCA1, and BRCA2. PARP inhibitors are also being evaluated in clinical trials.
The most commonly used assays for PARP activity employ biotinylated NAD+ as a substrate for PARP to incorporate biotinyl-ADP-ribose into the PAR chain on an immobilized histone substrate. The quantity of incorporated biotinyl-ADP-ribose is determined with streptavidin-HRP.
Disadvantages of this technique include:
1) Non-native NAD+ analogs as a substrate may yield different kinetics than unmodified NAD+.
2) A lack of flexibility in protein/peptide substrates
3) Multiple time-consuming wash steps.
To overcome these obstacles, EMD Millipore’s PARP1 Enzyme Activity Assay utilizes a fluorescently-coupled enzyme assay for Nicotinamide degradation to permit the use of native NAD+ substrate and flexibility with peptide/protein acceptor substrates.
Poly(ADP-ribose) (PAR) is a polymer of ADP-ribose present in the nucleus in a free state or as a post-translational modification on glutamate residues on a number of substrates. Synthesis and degradation of PAR are highly regulated, and its presence mediates DNA repair, chromatin structure, cell cycle, and cell death. A family of poly(ADP-ribose) polymerases (PARPs) catalyzes the addition of ADP-ribose moieties onto substrate proteins via cleavage of NAD+ and liberation of nicotinamide. In humans the PARP family is comprised of 17 members, and the members vary significantly in structure and biological function. PARP1 plays a key role in DNA damage repair, particularly base excision repair. It is activated by DNA strand breaks to add PAR to itself and to histones, which results in the recruitment of PAR-binding DNA repair factors, such as XRCC1, to the site of damage. PARP2 overlaps functionally with PARP1 in DNA damage repair, but PARP2 plays a distinct role from PARP1 in development of spermatogonia, adipose tissue, and thymus tissue. Other members of the PARP family are less characterized, and some may function as mono(ADP-ribosyl) transferases rather than polymerases. Consequently, a new nomenclature has been proposed that refers to the common ADP-ribosyltransferase activity of the members.
A number of PARP1 inhibitors have been developed to block repair of chemotherapeutic agent-induced genotoxic damage, and thereby potentiate the cytotoxic effect of chemotherapeutics in cancer patients. Inhibition of PARP1 appears to be particularly efficacious in conjunction with chemotherapeutics on cancer cells harboring mutations in components of homologous recombination DNA repair pathways, primarily BRCA1, and BRCA2. PARP inhibitors are also being evaluated in clinical trials.
The most commonly used assays for PARP activity employ biotinylated NAD+ as a substrate for PARP to incorporate biotinyl-ADP-ribose into the PAR chain on an immobilized histone substrate. The quantity of incorporated biotinyl-ADP-ribose is determined with streptavidin-HRP.
Disadvantages of this technique include:
1) Non-native NAD+ analogs as a substrate may yield different kinetics than unmodified NAD+.
2) A lack of flexibility in protein/peptide substrates
3) Multiple time-consuming wash steps.
To overcome these obstacles, EMD Millipore’s PARP1 Enzyme Activity Assay utilizes a fluorescently-coupled enzyme assay for Nicotinamide degradation to permit the use of native NAD+ substrate and flexibility with peptide/protein acceptor substrates.
Application
EMD Millipore′s PARP1 Enzyme Assay is a flexible and reliable, homogeneous, no-wash assay for quantifying PARP1 & PARP2 activity. Based upon a novel patent-pending technology, this easy-to-perform assay employs nicotinamidase to measure nicotinamide generated upon cleavage of NAD+ during PARP-mediated poly-ADP-ribosylation of a substrate (see figure 1), thereby providing a direct, positive signal assessment of the activity of PARP1 & PARP2.
To perform the assay, a PARP enzyme, β-NAD, activated DNA, test compound, and recombinant nicotinamidase enzyme are combined and incubated for 30 minutes. During the incubation, the activated DNA triggers PARP1 or PARP2 to produce poly(ADP-ribose) and nicotinamide. In a secondary reaction, the nicotinamidase enzyme converts the nicotinamide into nicotinic acid and NH3+ (free ammonia). To generate a signal for readout, a proprietary developer reagent is added and the signal is read using a fluorescent plate reader.
The robust performance of this assay makes it appropriate for measuring PARP1 and PARP2 activity as well as for screening of activators and inhibitors of PARP enzymes.
To perform the assay, a PARP enzyme, β-NAD, activated DNA, test compound, and recombinant nicotinamidase enzyme are combined and incubated for 30 minutes. During the incubation, the activated DNA triggers PARP1 or PARP2 to produce poly(ADP-ribose) and nicotinamide. In a secondary reaction, the nicotinamidase enzyme converts the nicotinamide into nicotinic acid and NH3+ (free ammonia). To generate a signal for readout, a proprietary developer reagent is added and the signal is read using a fluorescent plate reader.
The robust performance of this assay makes it appropriate for measuring PARP1 and PARP2 activity as well as for screening of activators and inhibitors of PARP enzymes.
Research Category
Apoptosis & Cancer
Apoptosis & Cancer
This PARP1 Enzyme Assay is a flexible & reliable, homogeneous, no-wash assay for quantifying PARP1 & PARP2 activity.
Components
Black 96 well plate - 1 plate
PARP1 Assay Buffer - 4 mL
Plate Covers - 2 plate covers
Activated DNA - 1 vial (100 µg at 10 mg/mL)
β-NAD - 1 vial (30 µL at 50 mM)
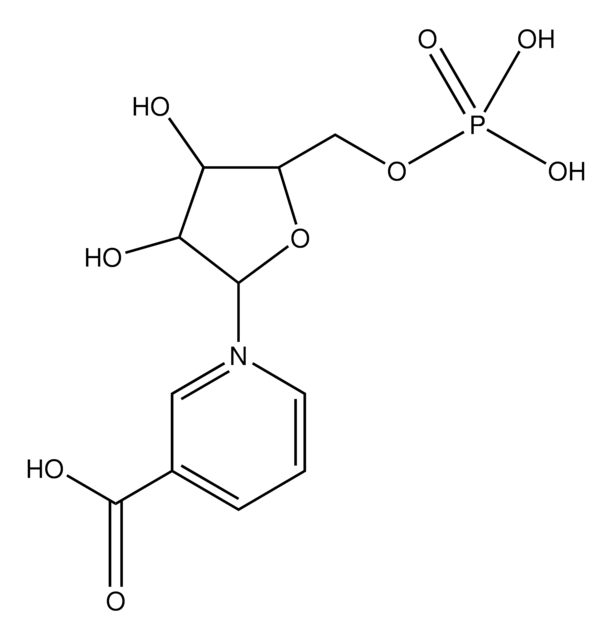
Nicotinamide (NAM) - 1 vial (10 µL at 10 mM)
Recombinant PARP1 Enzyme - 1 vial (2 µg at 0.2 µg/µL)
Recombinant Nicotinamidase Enzyme with 50% glycerol - 1 vial (60 µg at 1 mg/mL)
3-ABA (PARP Inhibitor) in DMSO - 1 vial (300 µL at 100 mM)
Developer Reagent - 4 mL at 10 mM.
PARP1 Assay Buffer - 4 mL
Plate Covers - 2 plate covers
Activated DNA - 1 vial (100 µg at 10 mg/mL)
β-NAD - 1 vial (30 µL at 50 mM)
Nicotinamide (NAM) - 1 vial (10 µL at 10 mM)
Recombinant PARP1 Enzyme - 1 vial (2 µg at 0.2 µg/µL)
Recombinant Nicotinamidase Enzyme with 50% glycerol - 1 vial (60 µg at 1 mg/mL)
3-ABA (PARP Inhibitor) in DMSO - 1 vial (300 µL at 100 mM)
Developer Reagent - 4 mL at 10 mM.
Storage and Stability
Maintain the unopened kit at -80ºC. Unused components should be stored at -80ºC. When stored properly this kit is stable for up to 4 months after date of receipt. Avoid repeated freeze/thaw cycles by aliquoting, if components are needed after first use.
Legal Information
CHEMICON is a registered trademark of Merck KGaA, Darmstadt, Germany
Disclaimer
Unless otherwise stated in our catalog or other company documentation accompanying the product(s), our products are intended for research use only and are not to be used for any other purpose, which includes but is not limited to, unauthorized commercial uses, in vitro diagnostic uses, ex vivo or in vivo therapeutic uses or any type of consumption or application to humans or animals.
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Flam. Liq. 3 - Repr. 1B
WGK
WGK 2
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Matteo Cassandri et al.
International journal of molecular sciences, 22(19) (2021-10-14)
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood. About 25% of RMS expresses fusion oncoproteins such as PAX3/PAX7-FOXO1 (fusion-positive, FP) while fusion-negative (FN)-RMS harbors RAS mutations. Radiotherapy (RT) plays a crucial role in local control but metastatic
Karin H Müller et al.
Cell reports, 27(11), 3124-3138 (2019-06-13)
Biomineralization of the extracellular matrix is an essential, regulated process. Inappropriate mineralization of bone and the vasculature has devastating effects on patient health, yet an integrated understanding of the chemical and cell biological processes that lead to mineral nucleation remains
H Yamaguchi et al.
Oncogene, 37(2), 208-217 (2017-09-20)
Inhibitors against poly (ADP-ribose) polymerase (PARP) are promising targeted agents currently used to treat BRCA-mutant ovarian cancer and are in clinical trials for other cancer types, including BRCA-mutant breast cancer. To enhance the clinical response to PARP inhibitors (PARPis), understanding
Patricia Santofimia-Castaño et al.
Communications biology, 5(1), 732-732 (2022-07-23)
Proteomic, cellular and biochemical analysis of the stress protein NUPR1 reveals that it binds to PARP1 into the nucleus and inhibits PARP1 activity in vitro. Mutations on residues Ala33 or Thr68 of NUPR1 or treatment with its inhibitor ZZW-115 inhibits
Hanlin Ma et al.
Cell death & disease, 13(3), 263-263 (2022-03-26)
Poly (ADP-ribose) polymerase (PARP) inhibitor (PARPi) resistance remains a therapeutic challenge in ovarian cancer. High-mobility group box 3 (HMGB3) plays significant roles in the development of drug resistance of many cancers. However, the function of HMGB3 in PARPi resistance is
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service
![[Met5]Enkephalin acetate salt hydrate ≥95.0% (HPLC), powder](/deepweb/assets/sigmaaldrich/product/structures/158/699/80b4b65b-7e48-49a2-ba61-4433cf1f375c/640/80b4b65b-7e48-49a2-ba61-4433cf1f375c.png)