Rapid Glycan Prep & Analysis by LC-MS Workflow
Overview
- We have developed a proprietary detergent-based buffer for rapid deglycosylation of N-linked glycans using PNGase F.
- Complete release of N-glycans is achieved in 15 minute incubation vs typical overnight digestions.
- Substrate reduction before PNGase F treatment was evaluated as an optional vs necessary step.
- Various molecular weight cut-off (MWCO) filters were tested for workflow compatibility.
- Required enzyme to substrate ratio was determined.
- Glycan profiles from PNGase Fast work-flow are comparable to those obtained with the longer, overnight protocols.
Products

Figure 1.Comparison of PNGase F workflows for glycan release. The traditional workflow (top) involves multiple wash steps and an overnight digestion. With PNGase Fast (bottom), digestion is complete in 15 minutes with minimal washes. Released glycans were labeled with procainamide by reductive amination and analyzed by LC-FL-MS.
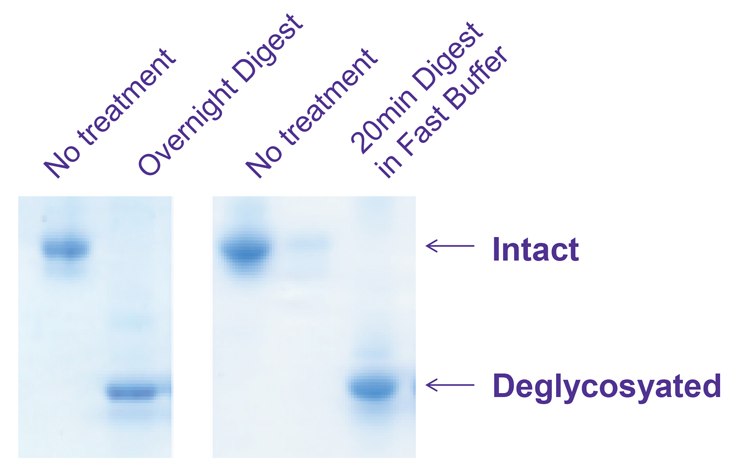
Figure 2.Left: Traditional overnight deglycosylation of bovine acid glycoprotein by PNGase F
Right: PNGase Fast deglyocosylation of bovine acid glycoprotein in 20 minutes
Bovine Acid glycoprotein (G3643) was deglycosylated with PNGase F using a conventional overnight protocol and our new fast protocol. Deglycosylated protein was recovered from the filter and analyzed by electrophoresis on a 4 - 20% Tricine gel and stained with EZBlue™ (G1041).
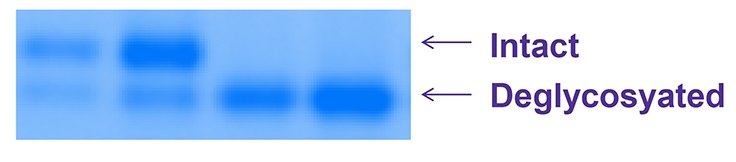
Figure 3.Deglycosylation of non-reduced (left two lanes) vs reduced (right two lanes) RNase B protein using Fast PNGase protocol. Reduction greatly enhances the deglycosylation of this protein.
RNAse B (R7884) was denatured with or without βME before deglycosylating using the fast protocol. Deglycosylated protein was recovered from the filter and analyzed by electrophoresis on a 4 - 20% Tricine gel and stained with EZBlue™ (G1041).
Glycan Distribution Overnight Digestion vs PNGase Fast
Cetuximab Glycan Distribution
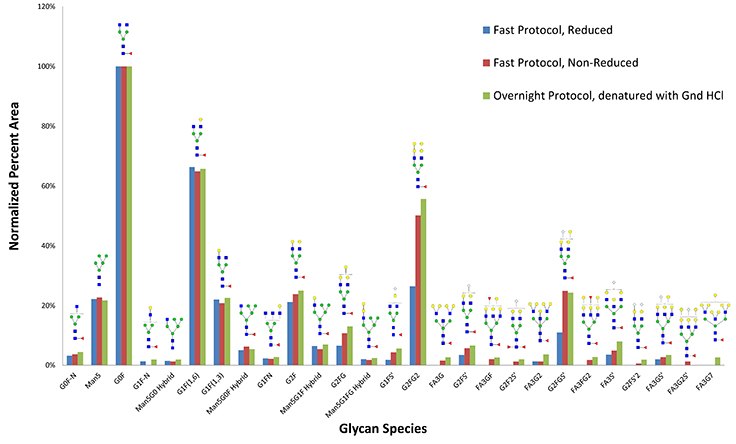
Figure 4.Glycan species recovered from reducing fast protocol (blue) compared to non-reducing protocol (red) and traditional overnight protocol (green). Reduction does not appear to be necessary for this monoclonal antibody.
Distribution of glycans recovered from Cetuximab monoclonal antibody when digesting with PNGase Fast protocol, with or without reduction, compared to conventional overnight digestion after denaturing with guanidine-HCl.
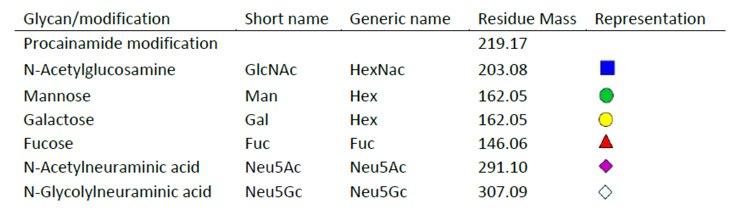
Figure 5.Legend of glycan sugars and corresponding masses used to determine overall glycan species putative structure.
Fast Protocol Filter and Enzyme Requirements
Monoclonal Human IgG1 SigmaMab
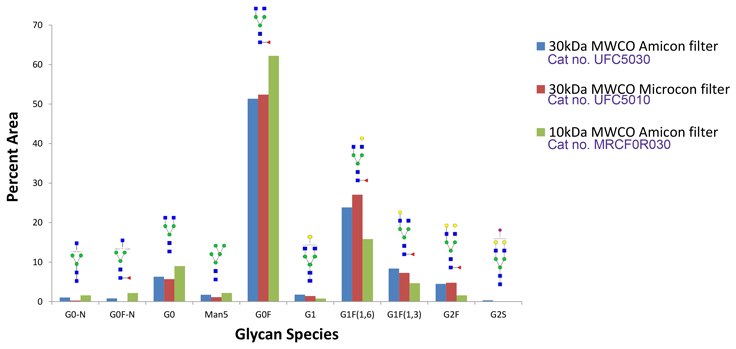
Figure 6.Glycan species recovered from fast protocol using 30kDa MWCO Amicon filter (blue) compared to glycans recovered from 30kDa MWCO Microcon filter (red) and 10kDa MWCO Amicon filter (green). Some changes in glycan profile are observed with the 10 kDa MWCO filter, therefore the 30 kDa filters are recommended.
SILu™Lite SigmaMAb universal human IgG standard (MSQC4) was deglycosylated with PNGase Fast using ultra filters with differing MWCO for detergent removal and glycan recovery.
IgG from human serum
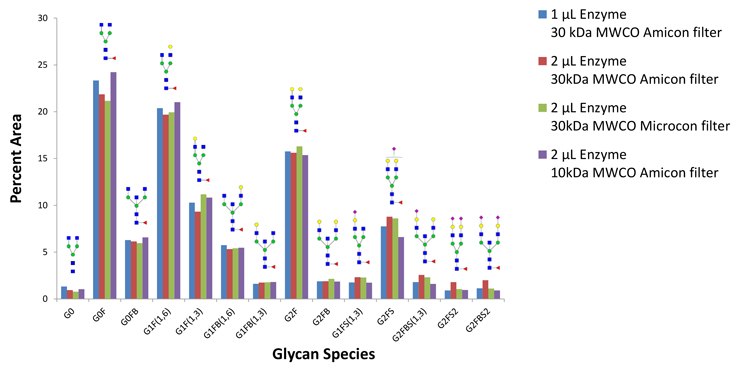
Figure 7.Glycans released by fast protocol and recovered from 30kDa MWCO Amicon filter using 1 µL of PNGase F (blue), compared to glycans recovered from 30kDa MWCO Amicon filter using 2 µL of PNGase F (red), glycans recovered using 30kDa MWCO Microcon filter using 2 µL of PNGase F (green), and glycans recovered from 10kDa MWCO Amicon filter using 2 µL of PNGase F (purple). In all cases, the recovered glycan profile is similar. PNGase Fast Kit includes 50 µL of PNGase F enzyme, sufficient for about 50 reactions with 100 µg glycoprotein.
100 µg IgG from Human serum (I4506) deglycosylated with PNGase Fast using different ultra filters and enzyme to substrate ratios.
Conclusion
- Use of PNGase Fast denaturing buffer and enzyme yielded results similar to a conventional 20-hour protocol with overnight digest while reducing workflow time to about 1 hour with a 15 minute digest.
- Substrate reduction prior to PNGase Fast treatment is optional, but required for complete deglycosylation of some samples.
- 10K and 30K MWCO filters performed comparably. 30K MWCO are recommended as they allow for shorter spin times and an overall faster workflow.
- 1 µL of enzyme was sufficient for full deglycosylation of 100 µg of human IgG.
- PNGase Fast buffer and enzyme can be used instead of conventional filter-assisted workflows to allow for comparable results with significant time savings.
To continue reading please sign in or create an account.
Don't Have An Account?