Ultrapure vs. DEPC Treated Water for Molecular Biology Applications
Nuclease-free glassware and reagents are required for many molecular biology applications. Chemical treatment with diethylpyrocarbonate (DEPC) has long been a common practice to generate nuclease-free water, but this method has several drawbacks. Alternatively, an ultrafiltration cartridge can be placed at the outlet of a water purification system in order to deliver nuclease-free ultrapure water that can directly be used for experiments.
Section Overview
Nuclease-free reagents for DNA and RNA research
Nucleases degrade DNA and RNA by cleaving the phosphodiester bonds of the nucleic acid backbone. While some nucleases are useful research tools, they are often a concern for scientists looking to preserve sample integrity for applications such as PCR, cloning, sequencing, or gene editing. For this reason, scientists working with nucleic acids require nuclease-free reagents and containers. Contamination by nucleases in DNA and RNA applications can lead to sample loss, lack of reproducibility, and experimental failure, making the use of nuclease-free materials essential for preparing samples, reagents, and buffers in such assays. Water, as the major component of buffers and reagents, should especially be nuclease-free. Unfortunately, nucleases are commonly found in the laboratory environment; they can be present on plasticware, in reagents, and even originate from the scientists through skin contact or saliva.1 The enzymes are very stable and difficult to inactivate;2 therefore, diethylpyrocarbonate (DEPC) treatment is often used to eliminate nucleases from solutions, including water.
Deionized water vs. nuclease-free water
Deionized (DI) water is purified water that has had its mineral ions—such as sodium, calcium, iron and copper—removed. However, DI water may still contain nucleases that can degrade nucleic acids. In contrast, nuclease-free water is specifically treated and tested to ensure it is free of RNases and DNases. Therefore, while all nuclease-free water is deionized, not all deionized water is free from nucleases.
Methods to prepare nuclease-free water
Nuclease-free water can be prepared using several methods, including chemical treatment with DEPC and ultrafiltration. DEPC treatment inactivates RNases, however, it can also introduce by-products that may interfere with experiments. Ultrafiltration, on the other hand, uses a membrane-based filtration process to remove nucleases, delivering RNase-free water on demand without the need for chemical treatments or autoclaving, thus ensuring high water quality and safety.
Challenges of Using DEPC-Treated Water in Molecular Biology
Nuclease contamination of reagents used in molecular biology, and particularly water, may lead to inconsistent results or loss of valuable samples. DEPC is an efficient, nonspecific inhibitor of RNase and has been used for many years to inactivate RNases.3 Chemical treatment of solutions with DEPC is an efficient method, but it presents several drawbacks:
- Safety concerns. DEPC is a suspected carcinogen, as it may react with purines, and should be handled with caution. In addition, if the bottle is exposed to traces of moisture from the air at room temperature, DEPC may hydrolyze. This will generate carbon dioxide, which will lead to potentially dangerous pressure buildup.4
- Time and energy consumption. Some time is required for the chemical reaction between DEPC and nucleases to occur, and then for autoclaving the solution, which is required in order to destroy the DEPC. In addition, autoclaving consumes large amounts of water and electricity.
- Introduction of impurities that may affect experiments. When DEPC is hydrolyzed during autoclaving, ethanol and carbon dioxide are released (Figure 1). Ethanol may react with impurities or reagents used in later experimental steps, potentially generating unwanted by-products. In addition, DEPC has a strong affinity for adenosine, and even if only traces of DEPC remain in solution, they may compromise experimental results by modifying adenosine nucleotides (Blots, PCR reactions, etc.).1 DEPC traces may also react with amine groups and mercaptans.
- Nucleases are not removed from the water, but merely inactivated by chemical reaction with DEPC.
- Limitation in PCR applications. DEPC and its hydrolysis products can lead to non-specific amplification or complete failure of the PCR reaction, compromising the integrity of the results.
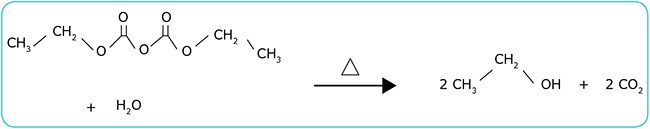
Figure 1.Degradation of DEPC releases contaminants into water.
Comparison: DEPC vs ultrafiltration
Ultrafiltration was tested as an alternative method to generate nuclease-free water. This process uses hollow fibers to separate contaminants based on their size. A disposable cartridge containing 13 000 Da nominal molecular weight limit (NMWL) cut-off polysulfone ultrafiltration fibers was tested (Figure 2).

Figure 2.Polysulfone hollow fiber used for ultrafiltration in the Biopak® cartridge.
Figure 3 shows that the rRNA remained intact when the RNase solution was first filtered using ultrafiltration. In comparison, conventional DEPC treatment of the RNase solution similarly prevented rRNA degradation. As a control, when the RNase solution was left untreated, rRNA was digested. This demonstrates that the efficiency of RNase removal by ultrafiltration is equivalent to the inactivation of RNase activity by DEPC treatment.5

Figure 3.Gel electrophoresis of rRNA in water previously spiked with RNase and either DEPC-treated, ultrafiltered, or left untreated.
Experiment: Effect of DEPC Treatment vs. Ultrafiltration on RNase Activity
Ultrapure water was spiked with RNase A and divided into three aliquots. The solutions were either (1) treated with DEPC and autoclaved; (2) treated with ultrafiltration through a Biopak® ultrafiltration polisher; (3) not treated. Ribosomal RNA was added to each solution, incubated for 20 minutes, and then agarose gel electrophoresis in denaturing conditions was performed.
Ultrafiltered Water: A Safer Alternative to DEPC Treated Water
Nuclease-free water is required for many molecular biology applications. Unlike DEPC treatment, which is time-consuming and cumbersome, using an ultrafiltration cartridge is a safe and convenient way to obtain nuclease-free water from a water purification system.
This method has many benefits for scientists:
- On demand, convenient. Freshly purified nuclease-free water can easily be produced when it is needed. There is no need to order bottled nuclease-free water in advance, or to spend time treating water with DEPC before performing experiments.
- No risk. No manipulation of chemicals is required. Also, since no chemicals are added to the water, there is no risk of interference with other reagents.
- High purity nuclease-free water. Ultrafiltration removes nucleases from water instead of merely inactivating them. Since the cartridge is designed to be connected to the outlet of a water purification system, such as a Milli-Q® system, one can obtain ultrapure water that is also free of nucleases.
For a further understanding of why ultrafiltered ultrapure water is preferable to DEPC-treated water, read the study “Water for Nuclease-sensitive Molecular Biology Studies”
Related Products
Acknowledgments
The authors are grateful to Julien Bole and Ichiro Kano for their technical expertise.
References
To continue reading please sign in or create an account.
Don't Have An Account?