Ultrapure Water for DNA Sequencing
In the rapidly evolving field of genomics, the speed and precision of DNA sequencing technologies are paramount. Contaminants from genomic DNA or reagents, including water, can significantly compromise outcomes. It is essential to use high-quality ultrapure water when preparing DNA libraries, sequencing samples, and reagents used in the process. This webpage explores the importance of high-quality purified water to optimize sequencing processes, ensuring the reliability and integrity of your genomic analyses.

What is DNA Sequencing?
DNA sequencing allows scientists to decipher the precise order of nucleotides present in a DNA fragment. This sequence gives insights into the genetic information encoded in the DNA and can help identify genes coding for proteins to gain a better understanding of the cell or organism. It can also uncover variations, mutations or genetic predispositions associated with diseases. DNA sequencing techniques have evolved over the years, becoming faster, more accurate and more cost-effective.1 They have a wide range of applications in fields such as medicine, agriculture, forensics and evolutionary biology.
Sanger and Second-generation Sequencing
Sanger sequencing was developed by Frederick Sanger in 1977. It involves amplifying DNA fragments using PCR, then incorporating chain-terminating dideoxynucleotides (ddNTPs) during DNA synthesis. The resulting fragments are separated by size using gel electrophoresis, and the sequence is determined by the position of the terminating nucleotide. Advancements of the technique include the incorporation of fluorescently-labeled nucleotides and automated computer analysis of nucleic acid fragments.
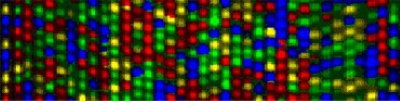
Figure 1.DNA sequencing lanes showing color-coded nucleotide bases.
Figure 1 shows a set of sequencing lanes where electrophoresis is used to separate molecules differing by one base. Laser detection is used to identify the bases at each position. The sequence is read from the bottom up, using a key where "A" is green, "C" is blue, "G" is yellow, and "T" is red. Software is used to determine the signal/noise ratios of the dyes for each position so that the proper base can be identified or "called".
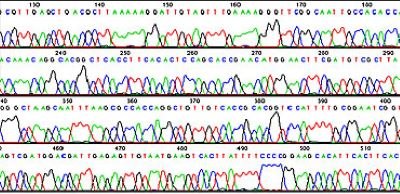
Figure 2.A software generated DNA sequencing chromatogram file showing the order of bases.
The order of the bases is displayed in a chromatogram or trace file as shown in Figure 2. A higher peak means a stronger signal, suggesting a more substantial presence of the detected nucleotide at that position. This can be due to a more efficient incorporation of the labeled nucleotide during the sequencing process. The height of the peak is also an indicator of the confidence in the base call.
Types of Advanced Sequencing Technology
There have been vast improvements in sequencing speed to accommodate the need for large scale genomic analysis. Next-generation sequencing (NGS) and third-generation sequencing encompass several modern high-throughput sequencing technologies that allow for parallel sequencing of millions of DNA fragments.2 Unlike Sanger sequencing, NGS doesn't require gel electrophoresis and uses various methods to simultaneously sequence many fragments, leading to faster and more cost-effective sequencing. These techniques allow the sequencing of whole genomes faster than ever and reveal complex patterns of gene expression that are revolutionizing genomic, transcriptomic, and proteomic analyses.
Ion Torrent Sequencing: Speed & Scalability
Ion Torrent sequencing detects pH changes caused by the release of hydrogen ions during DNA polymerization. It uses semiconductor technology to directly measure the ions released as each nucleotide is incorporated. This technique is known for its speed and scalability, making it suitable for various sequencing applications.
PacBio Sequencing: Long Reads & Structural Insights
PacBio sequencing utilizes single-molecule real-time (SMRT) sequencing technology. It involves observing DNA polymerase activity in real-time as it synthesizes DNA. Each nucleotide incorporation is detected by monitoring changes in the fluorescent signal of fluorescently labeled nucleotides. PacBio sequencing is renowned for generating long reads, enabling the sequencing of complex genomic regions and providing insights into structural variations.
Nanopore Sequencing: Portability & Real-Time Analysis
Nanopore sequencing is a disruptive technology that passes DNA strands through nanopores embedded in a membrane. As the DNA molecule translocates through the nanopore, it causes characteristic disruptions in the electrical current, which are used to identify the sequence of bases. This method offers advantages such as long reads, real-time sequencing, and portability, making it suitable for field applications and rapid sequencing.
In addition, NGS-based applications such as ChIP or single-cell sequencing provide a high-resolution, multidimensional approach to linking nucleic acid sequences to cell phenotypes.
Single-Cell RNA Sequencing: Insights into Gene Expression
Single-cell sequencing techniques enable the analysis of genetic material from individual cells. Various approaches exist, including single-cell RNA sequencing (scRNA-seq), single-cell DNA sequencing (scDNA-seq), and single-cell ATAC-seq (scATAC-seq). These techniques provide insights into cellular heterogeneity, cell lineage, and the genetic basis of diseases at a resolution not possible with bulk sequencing methods. They involve isolating single cells, amplifying their genetic material, and sequencing it using NGS technologies adapted for low-input samples.
Impact of Water Quality on DNA Sequencing
The speed and precision of advanced DNA sequencing makes it imperative to avoid impurities or genomic contamination. The quality of the purified water used can affect the accuracy of the results. Water quality used to prepare DNA samples and libraries, reagents, and operate the sequencer should meet stringent quality criteria to optimize the sequencing process.
Water contaminants may impact sequencing outcomes as follows:
Nucleases and DNA Stability
Nuclease-free water is recommended to avoid the degradation of the DNA at all steps of the DNA sequencing workflow, regardless of the method used. Nuclease removal is efficiently done by ultrafiltration. Point-of-use ultrafiltration cartridges (such as the Biopak® polisher) can be installed at the outlet of water purification systems to provide nuclease-free water on demand.
Organic Contaminants
Organic molecules are the most disruptive contaminants in DNA sequencing. Large organic acids, such as humic and fulvic acids resulting from natural matter degradation can co-elute with the DNA fragments during electrophoresis and can disturb the polymerase efficiency as well during the PCR step.
In addition, and more frequently, some organics can interfere with fluorescence detection and contribute to errors in the sequencing of the target DNA. The presence of organic molecules absorbing and quenching fluorescence can disturb its detection. Many sequencing techniques are based on the detection of fluorescent dyes attached to the DNA fragments (Illumina, PacBio, Sanger, etc.) and may be affected by this. To ensure organic purity of the high purity water used for DNA sequencing, a low overall organic content, referred to as Total Organic Carbon (TOC), is recommended.
Ions
Ions can have an impact on several processes used in various sequencing technologies. The ionic strength needed during the electrophoresis process is best obtained and controlled when the water used has a high purity and does not contain unknown concentrations of ions. The levels of some ions, such as magnesium, need to be carefully adjusted during PCR, to optimize the efficiency of the polymerase.
Other ions, such as cadmium and some other divalent cations, must be removed to prevent inhibition of the polymerase. Impurities in laboratory water can affect the stability and conductivity of the electrolyte solution used in nanopore sequencing, leading to fluctuations in the electrical signal and reduced sequencing accuracy. Finally, ion levels are especially important for Ion Torrent sequencing, which is based on the detection of pH changes.3 The water used should contain as few ions as possible, so there is no influence on the pH.
High resistivity (18.2 MΩ•cm) ensures low ionic content (below 1 µg/L of overall ionic concentration) in water. High purity (ultrapure) water purification systems are designed to provide continuously high resistivity water on demand.
Bacterial Contamination
Bacteria can release nucleases in the water and therefore should be removed to avoid degradation of the nucleic acids. Bacteria also release DNA that could interfere with the electrophoresis separation of the target DNA fragments. In addition, bacteria release ions and organics, which are contaminants that can interfere with the DNA sequencing, as mentioned previously. Membrane technologies, absolute 0.2 µm microfiltration or ultrafiltration, can remove bacteria at the point of water delivery and ensure low bacteria levels.
In summary, impurities in laboratory water can impact DNA sequencing techniques by affecting the accuracy, reliability and reproducibility of sequencing data. Therefore, it's crucial to use high-quality water and regularly monitor water purity to ensure optimal performance in DNA sequencing experiments.
Importance of Type 1 Water in Genomic and DNA Sequencing
In conclusion, ultrapure (Type 1) water purified with an ultrafiltration final filter is highly desirable for genomic sequencing and related techniques to avoid contamination and improve repeatability.
A variety of water purification solutions tailored for DNA and RNA library work are available. Advanced water purification systems that use ultrafiltration cartridges, such as the Biopak® polisher, can provide nuclease-free water on demand, further reducing contamination risks.
Investing in a high-quality water purification system is vital for precision in genomic research. Milli-Q® ultrapure water supports researchers in obtaining reliable sequencing data, facilitating groundbreaking discoveries in genomics, transcriptomics, and proteomics.
Related Products
References
To continue reading please sign in or create an account.
Don't Have An Account?