Identification of Cell Culture Media with Raman Spectroscopy
Abstract and Introduction
Identification of cell culture media samples by traditional LC methods, such as amino acid or vitamin analysis, has high costs, and requires significant analytical expertise and laboratory space. Raman spectroscopy offers many potential benefits, such as low cost, portability, and potentially limited skill required to operate the instruments (Figure 1 for a brief introduction to Raman spectroscopy).
A comprehensive analysis of raw material spectral contributions to dry powder media spectra was performed in order to evaluate the capabilities of Raman spectroscopy for dry powder media identification and raw material quantitation. Spectra of pure solid raw materials were obtained and used for classical least squares (CLS) analysis of chemically defined dry powder media spectra.
The analysis reveals that sensitivity to low mass percent raw materials and raw material polymorphism limits the capability of Raman spectroscopy for identification of complex chemically defined dry powder media. Using the predictive power of the CLS model, simple feeds where Raman spectroscopy was predicted to be an adequate quantitation and identification analysis tool were selected. Data supporting the application of a CLS model and the key learns obtained from application of the CLS model will be discussed.
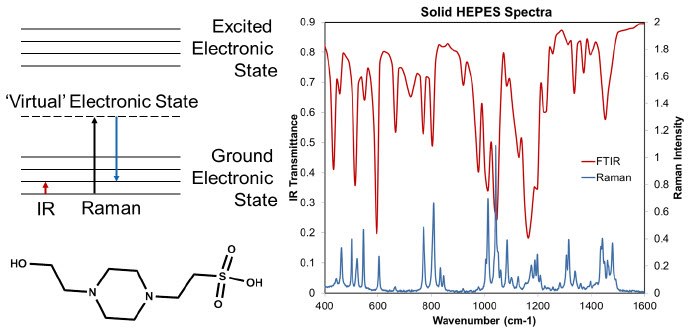
Figure 1.On the left the chemical structure of HEPES, and an energy diagram showing the differences between Raman and IR are shown. On the right FTIR and FT-Raman spectra of HEPES are shown. FTIR is a direct vibrational absorbance technique, and Raman an indirect measure of the vibrational spectrum. In Raman spectroscopy the sample is exposed to monochromatic light, and some of the scattered light is shifted to a lower energy because molecules in the sample have been promoted to a higher vibrational state. The Raman scattering is typically stronger for higher energy light, but with higher energy light there is also typically more background fluorescence. On the right, some of the same vibrational bands are observed, but there are different ratios of intensity for the bands.
Data Collection and Analysis
Solid samples of raw materials (amino acids, vitamins, polymers, etc.) and formulations (complete media and feeds) were analyzed by placing the solid samples into vials or tubes and placing them in the 1064 nm laser beam from a Bruker FT-Raman MultiRAM instrument. The laser power was 500 mW, resolution was 2 cm-1, and 128 scans were obtained. The formulations were rotated during analysis to cover more surface area and obtain more reproducible spectra1.
Prior to analysis by CLS the data was truncated (700-1800 cm-1), the first derivative was obtained, and the data was smoothed using a Savitsky-Golay algorithm2. The spectra from the raw materials and the spectra from the formulations were used to predict the mass percent of each raw material using a CLS model (Figure 2).
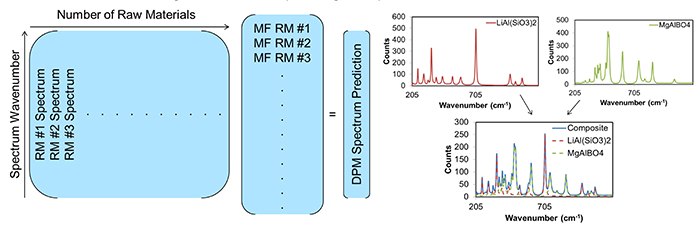
Figure 2. The CLS model is shown on the left, and an example of using a CLS model to generate a spectral prediction is shown on the right. On the left, raw material spectra (RM #x spectrum) can be multiplied by their respective mass fractions (MF RM #x) to generate a prediction for the dry powder media spectrum (DPM spectrum prediction). The spectra on the right show an example of summing up the individual spectrum to generate a spectrum of the mixture.
Sensitivity to Raw Materials in Dry Powder Media (DPM)
The CLS model from Figure 2 was used to fit the DPM spectra and obtain the spectral contributions of each raw material to the DPM spectrum. Figure 3 shows both the mass percent of each raw material, and the percent contribution of each raw material to the Raman spectrum. Contrasting the two pie charts reveals the lack of sensitivity of Raman to materials below 1-2%.

Figure 3. The pie chart on the left shows raw material mass percent contirbutions in the DPM formulation. The pie chart on the right shows the increase in spectral percent match by including that raw material spectrum in the DPM spectrum fit. When only L-Asparagine (L-Asp) and dextrose raw material spectra are used to fit the final DPM spectrum, there is a 97% match to the final DPM spectrum.
Raw Material Polymorphism
Polymorphism is the ability of neat chemicals to exist in more than one crystalline structure. When raw materials have different crystalline structures, they have different vibrational spectra (Figure 4). This difference in vibrational spectra of raw materials for the same primary chemical structure can lead to false negatives when distinguishing between DPM spectra with percent matching or principal component analysis (PCA).
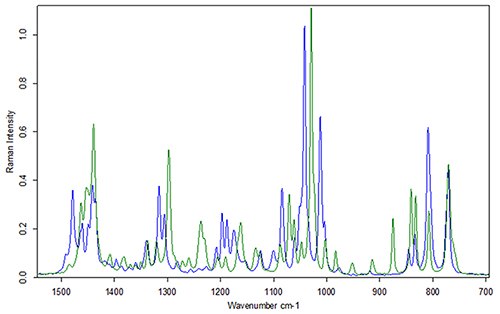
Figure 4. Raman spectra of HEPES from two different vendors. The primary chemical structure is the same (LC/MS not shown), and the Raman spectra are identical when dissolved in water (Raman spectra not shown).
Comparison of DPM Spectra by PCA
Six different media products were evaluated in this study. Medium 1 is a feed, media 2-4 and 6 are complete media, and medium 5 is a concentrated medium. Media 3 and 6 are similar, but medium 6 has twice the dextrose. Media 2 and 4 are similar, but medium 4 has a slight increase in dextrose and HEPES.

Figure 5. PCA scores plot of the DPM lot spectra from all media evaluated. The ovals are the 95% confidence intervals for that particular medium. Media 3 and 6 have very large confidence intervals because of polymorphism. Media 2 and 4 are indistinguishable because of the lack of sensitivity to differences in low mass percent raw materials.
Conclusions
Raman spectroscopy for identifying low level impurities in raw materials is not reccomended due to low sensitivity. Raman spectroscopy for unique identification of dry powder media should be limited to products that have difference in high mass percent Raman active components. Polymorphism of high mass percent Raman active materials must be addressed in modelling.
References
To continue reading please sign in or create an account.
Don't Have An Account?