Rapid Trypsin Digestion of Complex Protein Mixtures for Proteomics Analysis
Abstract
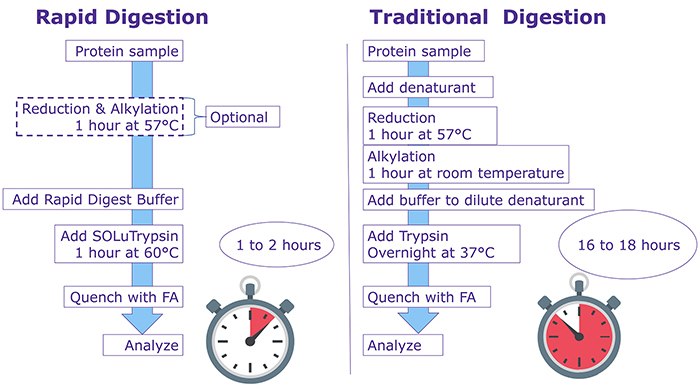
Reliable and reproducible results in mass spectrometry proteomics analyses are highly dependent on robust sample preparation. Sample preparation workflows are often cumbersome and generally involve chemical denaturation, reduction and alkylation, buffer exchange, and overnight protease digestion of the protein sample. These lengthy, complex protocols require 3-4 hours of lab work followed by overnight incubation, potentially introducing variability to results. Filter-assisted sample prep (FASP) and other alternative protocols can speed up the workflow but with increased costs and limited applicability. In this study, we identified conditions allowing for rapid trypsin digest at elevated temperatures that yielded reliable, reproducible results in less than 2 hours on a wide variety of substrates.
Workflow
All reagents are from MilliporeSigma. TCEP (646547), CAA (C0267), IAA (A3221), Sodium deoxycholate (30970),
Rapid Digest Buffer (component of Rapid Digest Kit, MSKT0002), SOLu-Trypsin (EMS0004), Formic acid (695076),
bovine serum albumin (P0914), myoglobin (M1882), Amicon Ultra (UFC5030). K562 and CHO were produced in house.
Results: Purified Proteins
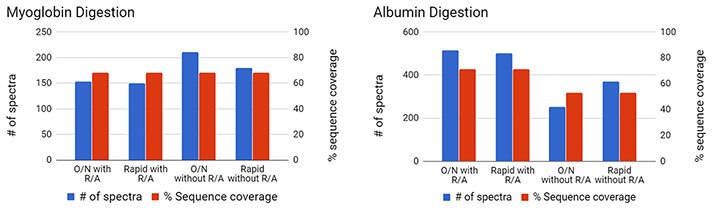
Figure 1: Digestion of Purified ProteinsDigest of individual proteins using either a traditional overnight (O/N) or rapid protocol, with or without reduction and alkylation (R/A). Database parameters utilized include no missed cleavage and 80% confidence for peptide. Left: Myoglobin, no cysteines Right: Serum Albumin, 6% cysteine
Rapid digest of individual proteins yielded comparable sequence coverage compared to overnight digestion. Proteins with high cysteine content require reduction and alkylation for efficient digestions.
Results: Complex Proteins – CHO
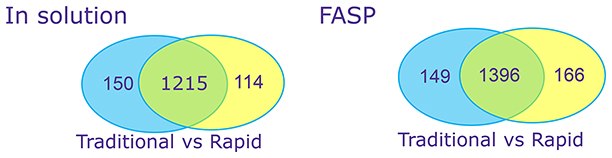
Figure 2: Complex Proteins – CHO host cell proteinsTraditional versus rapid digest of Chinese Hamster Ovary (CHO) secreted proteins, with and without Filter-Assisted Sample Preparation (FASP). Proteins were denatured, reduced and alkylated in traditional digests but not in rapid digests.
Rapid digestion yielded similar numbers of peptides and proteins as a traditional overnight digestion without the need for denaturing or R/A.
Results: Complex Proteins – K562 Lysate
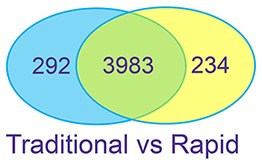
Figure 3: Complex Proteins – K562 Cell Lysate K562 cell lysates were prepared with SDS. Urea is used, with FASP, to remove the detergent prior to digestion. Proteins were digested using the traditional overnight workflow with R/A or the rapid workflow without R/A.
The two digestion methods result in similar numbers of peptide and protein IDs. The digests give consistent results in replicate preparations.
Good correlation is observed between the two methods for the proteins identified.
Compared to traditional protocols, the rapid digest approach identified similar numbers of peptides and proteins in complex mixtures, without the need for chemical denaturation or reduction and alkylation. Overall, these results suggest that incorporation of rapid digest buffer and elevated temperature can reduce the labor requirements and turnaround time of sample preparation for proteomics analyses by mass spectrometry.
Conclusions
- Digestion of complex protein samples in 2 hours with rapid digestion buffer and increased temperature gives results similar to traditional overnight digestion
- Traditional and rapid digestion produce similar numbers of peptide and protein identifications, and similar sequence coverage
- Rapid digestion is compatible with reduction and alkylation, which may be required for proteins high in cysteine
- Reduction and alkylation is optional for complex proteomics samples
Materials
To continue reading please sign in or create an account.
Don't Have An Account?