Dual-Labeled Probes
Dual-Labeled Probes are the most common probe type for qPCR. They are linear hydrolysis probes used in the 5’ nuclease assay with 5' reporter dyes and quenchers on the 3' end, in an internal position (between bases 9 and 10), an internal position (between bases 9 and 10) and the 3' end, or MGB linked to EDQ on the 3' end, respectively.
Read more about
Applications of Dual-Labeled Probes
- Microarray validation
- Multiply target genes / few samples
- Pathogen detection
- Multiplexing
- Viral load quantification
- Gene expression analysis
- Gene copy determination
Perfect for validating MISSION® siRNA and shRNA knockdowns.
BENEFITS OF USING DUAL-LABELED PROBES
- Design simplicity for sequence specificity
- Extensive availability of reporter/quencher combinations
- Increased sensitivity
ADD LOCKED NUCLEIC ACID OR MGB:EDQ TO YOUR PROBE
Benefits of Locked Nucleic Acid with BHQ™
- Increased thermal stability and hybridization specificity
- Greater accuracy in SNP detection, allele discrimination, and in vitro quantification or detection
- Easier and more sensitive probe designs for problematic target sequences
- Has no native fluorescence (emits heat instead of light), resulting in lower background fluorescence
- Increased signal-to-noise ratio, providing higher sensitivity
- Enables a wider choice of reporters for multiplexing
Benefits of MGB:EDQ
- Increased specificity of the probe-target hybridization, reducing the likelihood of non-specific binding
- Higher sensitivity enables the use of shorter probes, which increases the sensitivity of qPCR assays
- Melting temperature adjustment allows for Tm adjustment of the probe, which can be beneficial for designing qPCR probes with optimal Tm values
- Reduced background signal contributes to a reduction in background fluorescence, leading to an improved S:N or signal-to-noise ratio
- Enhanced stability can improve the stability of the probe-target hybrid, reducing the likelihood of probe degradation
- Design flexibility expands the options for probe design, facilitating the creation of efficient qPCR assays
- Enhanced SNP detection via mismatch discrimination
How Dual-Labeled Probes Work
A Dual-Labeled Probe is a single-stranded oligonucleotide labeled with two different dyes. A reporter dye is located at the 5’ end and a quencher molecule located at the 3’ end. The quencher molecule inhibits the natural fluorescence emission of the reporter by fluorescence resonance energy transfer (FRET). The illustration below depicts the mechanism.
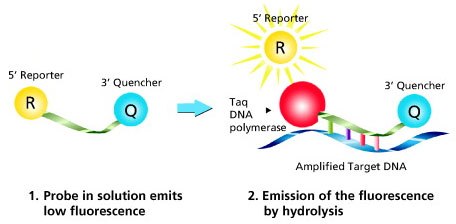
The primer is elongated by the polymerase and the probe binds to the specific DNA template. Hydrolysis releases the reporter from the probe/target hybrid, causing an increase in fluorescence. The measured fluorescence signal is directly proportional to the amount of target DNA.
Features of Dual-Labeled Probes
- Amounts: 1, 3, 5, and 10 OD
- Purification: HPLC
- Sequence Lengths: 15 - 40 bases
- Quality Control: 100% mass spectrometry
- Format: Supplied dry in amber tubes
- Custom formats available (normalization, special plates, etc.)
Our probes are provided in a format to simplify your experimental planning.
1JOE/TET alternative
2VIC® alternative
3Cyanine 3 alternative
4TAMRA™ alternative
5Cyanine 5 alternative
To continue reading please sign in or create an account.
Don't Have An Account?