Oligonucleotide Handling & Stability
DNA Oligonucleotides are relatively stable molecules (Table 1). However, they require certain handling and storage techniques to ensure trouble-free experiments and maximize shelf life, respectively.
Table 1. Approximate shelf-life of non-modified, single-stranded DNA oligonucleotides with proper storage. These values are estimates and are not guaranteed.
Resuspension, Storage, and Working Habits
Non-modified DNA
Resuspension. Unless requested to ship in solution, all single-stranded DNA oligonucleotides ship dry and are ready for use upon resuspension with the exception of thiol-modified sequences (follow this reduction protocol to activate these oligonucleotides) . Weak buffers, such as TE (10 mM Tris, pH 7.5 to 8.0, 1 mM EDTA) or Tris (10 mM Tris-HCl, pH 8.0) are preferred, with TE as the best choice. If TE is unsuitable for the end technique / application, then sterile, nuclease-free water is the second best choice.
Storage. As shown in Table 1, storage at -20 °C will provide the longest shelf life. TE buffer is the absolute best option as laboratory-grade water is often slightly acidic, which slowly degrades DNA. In addition, dry oligos are never truly dry. Some moisture always remains, which in turn leads to slow degradation. However, there is unlikely to be any significant difference in stability for at least 2 years whether stored dry, in water, or in buffer.
Non-Modified RNA
Resuspension. Single-stranded RNA oligonucleotides should be resuspended as described above for non-modified DNA.
Storage. RNA is much more unstable than DNA due to its chemical structure. RNA is subject to autocatalysis (Figure 1) as well as degradation by RNases, which are abundant in laboratories due to their presence in sources such as sloughed-off skin cells and ubiquitous microorganisms.
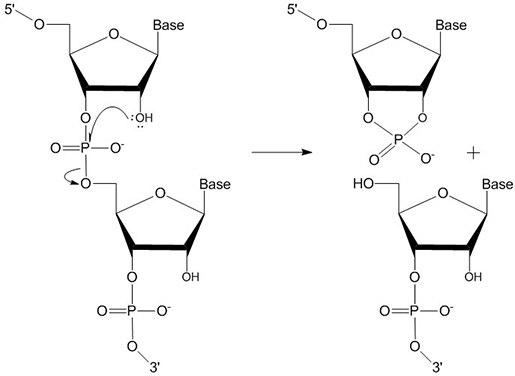
Figure 1. RNA autocatalysis.The 2’-hydroxyl group of RNA can lead to autocatalytic degradation, which in turn, under the right conditions, lowers the quantity of useful oligonucleotide.
For short-term storage, single-stranded RNA oligonucleotides should be stored in TE buffer at -80 °C. For the long-term, storage at -80 °C as an ethanol precipitate is the best option. Taking precautions to minimize exposure to RNases will increase shelf life.
Working with RNA. RNases often contain numerous disulfide bonds and therefore are very stable. Short of using harsh chemicals to eliminate these enzymes, they are resistant to common methods of decontamination, such autoclaving. The following precautions will help to minimize your RNA oligonucleotide to RNase exposure:
- Assume everything in your workspace / laboratory is contaminated with RNases.
- Use an isolated part of the lab that is dedicated to working only with RNA.
- Routinely clean the RNA-isolation area with agents that destroy RNases.
- Use chemicals, reagents, and water that are certified as RNase free.
- Use tools, such as micropipettes, that are for working only with RNA.
- Always wear gloves and change them after touching other surfaces.
Modified Oligonucleotides
Resuspension. Modified oligonucleotides, whether DNA or RNA, should be resuspended in TE buffer. This is critical, especially for fluorescent moieties, because pH can have a big impact on the intensity of fluorescent emission.
Storage. Modified oligonucleotides should have stability profiles similar to those of their unmodified DNA and RNA counterparts. Therefore, they should be stored according to the above recommendations for DNA and RNA. To prevent photobleaching, fluorescently-labeled oligonucleotides should be stored in the dark.
Working with Fluorescent Modifications. To prevent photobleaching when working with fluorescent moieties, keep them in their amber tubes or if transferred to clear tubes, wrap them in foil.
Duplex Oligonucleotides
Our scientists have found siRNA duplexes to be stable for at least 3 years when stored dry at -20 °C. DNA duplexes should be just as stable, if not more so (This value is an estimate and is not guaranteed. To receive an experimentally-determined expiration date with a guarantee, please inquire about a stability study [below] with us).
Single-Use Aliquots
While freeze / thaw cycles are believed to degrade genomic DNA, likely via shearing due to ice crystal formation1, our scientists have found that oligonucleotides are not negatively impacted by such cycles. However, dispensing oligonucleotides into single-use aliquots and then freezing them at -20 °C is still best practice. This prevents waste due to mishaps such as cross contamination or spills. For example, if you spill your single tube of oligonucleotide on the floor, you will have to reorder it. However, if you spill a single aliquot on the floor, you can simply thaw another tube.
Preparing Solutions
Oligonucleotide quantity, found on both the tube label and technical datasheet, is provided in a variety of units, including optical density (OD), micrograms (µg; by extension, µg/OD), and nanomoles (nmol). Our quantification method begins with taking an absorbance reading at 260 nm in a UV/Vis spectrophotometer to provide an OD value—all other units are derived from this OD value (to learn more, see Oligonucleotide Quantification). These units of quantity—nmol is the most useful—can be used directly to create stock solutions.
There are many units of concentration, but micromolar (µM) is probably the one most commonly used with oligonucleotides. Here, beginning with the already determined nmol quantity found on the tube label / technical datasheet, both longform and shortcut calculations needed to create a 100 µM stock solution as well as a 10 µM working solution will be provided as an example. For quicker results, including calculation of concentration in other units, please use the resuspension and dilution modules of OligoEvaluator™.
Example Longform Resuspension Calculation
We prefer to work in base units to help prevent ‘conversion confusion’ during dimensional analysis. Making mistakes is easy to do when directly trying to convert between nmol, µM, and other sub-base units.
Step 1. Convert the number of nmol on the tube label / technical datasheet to moles.
Our hypothetical oligonucleotide arrives with a quantity of 49.9 nmol.
mol = 49.9 nmol x 1 mol 109 nmol-1
mol = 4.99 x 10-8
Step 2. Convert the desired stock solution concentration of 100 µM to molarity.
100 µM = 100 µmol L-1
M (mol L-1) = 100 µmol L-1 x 1 mol 106 mol-1
M (mol L-1) = 1 x 10-4
Step 3. Use the desired stock solution concentration in molarity and the number of moles to calculate the volume in liters needed for resuspension.
1 x 10-4 mol L-1 = 4.99 x 10-8mol
L = 4.99 x 10-8mol / 1 x 10-4mol
L = 4.99 x 10-4
Step 4. Convert liters to microliters (µL).
Volume = 4.99 x 10-4 L x 106 µL L-1
Volume = 4.99 µL
Example Shortcut Resuspension Calculation
Step 1. Take the number of nmol from the tube label / technical datasheet and multiply by 10 to get the resuspension volume in microliters.
Volume = 4.99 nmol x 10
Volume = 499 µL
This shortcut calculation only works for creating 100 µM solutions.
In this particular case, add 499 µL of water or buffer to the tube containing the oligonucleotide and then vortex to ensure adequate mixing.
Additional Thoughts on Resuspension
This above resuspension example used 100 µM because it is a typical laboratory stock solution concentration. Creating stock solutions with concentrations too far below 100 µM can be problematic because the volume needed for resuspension can easily exceed the 2 mL capacity of the tube. For example, to create a 20 µM stock solution, this particular oligonucleotide, which is present at 49.9 nmol, would need to have 2.49 mL of water or buffer added to it.
Likewise, trying to create a stock solution more concentrated that 100 µM can also be problematic. For example, to create a 1000 µM stock solution, this particular oligonucleotide would need 49.9 µL of water or buffer. This small volume can be challenging to pipette accurately from a 2 mL tube.
To create a 100 µM stock solution, calculations and online tools are actually unnecessary as the volume needed for resuspension is included on the technical datasheet.
Example Working Solution Calculation
In this example, we will assume that the final 10 µM working solution is to be in a volume of 20 µL, which is typical for PCR.
Step 1. Use this concentration, volume equivalence formula.
C1V1 = C2V2
100 µM x V1 = 10 µM x 20 µL
V1 = 2 µL
In this particular case, add 2 µL of the stock solution to the reaction tube, and along with the other components, bring the final volume with water or buffer to 20 µL.
Quantity Verification
We take great care to ensure that our OD values are accurate. However, you may still want to verify the quantity of your oligonucleotide. In this case, if you have a traditional UV/Vis spectrophotometer, the method is simple.
Follow these steps:
- Resuspend the oligonucleotide in 400 µL of water or buffer.
- Dilute 12 µL into 988 µL of sterile, nuclease-free water.
- Take an A260 reading of the 1 mL sample in a cuvette.
- Ensure the OD value is in the linear range (~0.1 to 1 OD).
- Multiply the OD of the sample volume by the dilution factor.
Total OD = OD of 1 mL sample X 400/12 - Multiple total OD by µg/OD (from technical datasheet) to get µg.
References
To continue reading please sign in or create an account.
Don't Have An Account?