Performing a Purification and On-column Refolding of an Insoluble Histidine-tagged Protein
This procedure uses a HisTrap™ FF 1 mL column but can also be used with a HisTrap™ HP 1 mL or a HisTrap™ FF crude 1 mL column. The procedure uses ÄKTAprime plus but can also be run on other ÄKTA systems.
Alternative binding buffers: 5 to 40 mM imidazole can be included in the binding buffer to reduce unwanted binding of non-histidine-tagged proteins. The concentration of imidazole is protein dependent, and if the protein of interest elutes or does not bind at a certain imidazole concentration, reduce the concentration.
DISRUPTION, WASH, AND ISOLATION OF INCLUSION BODIES
- Resuspend the cell paste from 100 mL culture in 4 mL resuspension buffer.
- Disrupt cells with sonication on ice (e.g., 4 × 10 s).
- Centrifuge at 10 000 × g for 10 min at 4 °C.
- Remove supernatant and resuspend pellet in 3 mL of cold isolation buffer. Sonicate as above.
- Centrifuge at high speed for 10 min at 4 °C.
- Repeat steps 4 and 5.
At this stage the pellet material can be washed once in buffer lacking urea and stored frozen for later processing.
SOLUBILIZATION AND SAMPLE PREPARATION
- Resuspend the inclusion body pellet in 5 mL of binding buffer.
- Stir for 30 to 60 min at room temperature.
- Centrifuge for 15 min at high speed, 4 °C.
- Remove any remaining particles by passing the sample through a 0.45 µm filter.
The optimal concentration of 2-mercaptoethanol (0 to 20 mM) must be determined experimentally for each individual protein.
If it has not been prepared as above, adjust the sample to the composition of binding buffer by diluting in binding buffer or by buffer exchange using HiTrap Desalting or HiPrep 26/10 Desalting, then pass the sample through a 0.45 µm filter.
PREPARING THE SYSTEM
The requirement for linear gradient formation for refolding and elution makes the use of a chromatography system essential.
This example uses ÄKTAprime plus. Once the system is prepared, the remaining steps will be performed automatically. A model chromatogram is shown in Figure 1.
- Place each inlet tubing from port A (8-port valve) in eluents as given above and the tubing from port B (2-port valve) in the elution buffer.
- Place the three brown waste tubings in waste.
- Connect the column between port 1 on the injection valve (7-port valve) and the UV flow cell.
- Fill the fraction collector rack with 18-mm tubes (minimum 40) and position the white plate on the fractionation arm against the first tube.
- Connect a sample loop large enough for your sample between port 2 and 6 on the injection valve. Use a syringe to manually fill the loop.
Note: If a Superloop is needed, additional information is supplied in the instructions for Superloop.
SELECTING APPLICATION TEMPLATE AND STARTING THE METHOD
- Check the communication to PrimeViewTM. At the lower right corner of the screen the text Controlled By: Prime should be displayed.
- Use the arrow and OK buttons to move in the menu tree until you find On-Column Refolding HisTrap™.
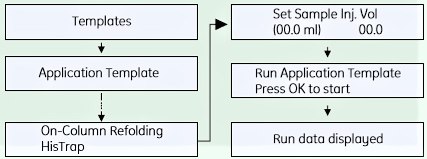
- Enter the sample volume and press OK to start the template.

Figure 1.Results for on-column refolding of a histidine-tagged protein. For conditions, see the protocol above. Refolding of histidine-tagged membrane proteins from inclusion bodies using IMAC has also been reported.
1The columns are prepacked with Ni Sepharose 6 Fast Flow (FF) or Ni Sepharose High Performance (HP) which are precharged with Ni2+ ions.
SCREENING CONDITIONS FOR REFOLDING USING IMAC
To screen for suitable IMAC refolding conditions, a 96-well microplate prepacked with Ni Sepharose can be used (Table 2). An example is shown in Figure 2.
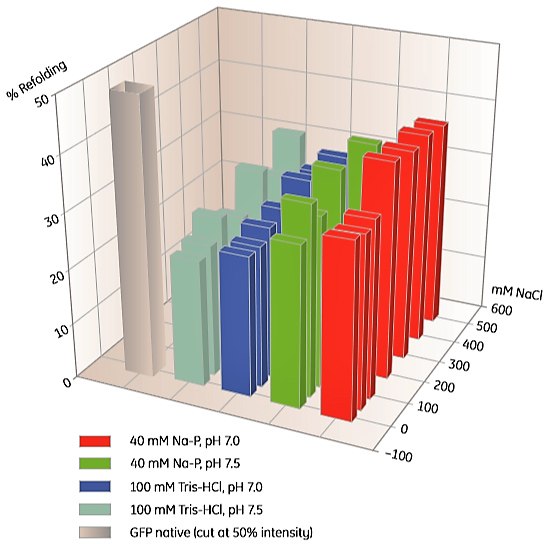
Figure 2. Screening of IMAC refolding conditions for histidine-tagged GFP using His MultiTrap FF. This initial screen covered buffer substances, pH and salt concentrations. Data kindly provided by J. Buchner, M. Haslbeck and T. Dashivets, Munich Technical University, Germany.
Materials
如要继续阅读,请登录或创建帐户。
暂无帐户?