Biosimilar Comparability Workflow for Intact Mass Analysis Using Trastuzumab as Model Protein
Introduction
Biosimilar mAb drugs have surged in the last decade due to patent expiration of innovator drugs, offering affordable care to patients. Biosimilars, while sharing amino acid sequences with innovators, can differ structurally due to post-translational modifications (PTMs) resulting from changes in manufacturing processes. These variations, like N-glycosylation and alterations to termini, pose challenges in proving comparability.1
Thorough characterization of therapeutic mAbs is vital for safety and efficacy and establishing critical quality attributes (CQAs) are essential for both innovator and biosimilar drugs.2,3 Characterization generally involves chromatography (e.g., SEC, RP, HILIC) combined with mass spectrometry (MS).
Trastuzumab received approval for medical usage in the United States in September 1998 and in the European Union in August 2000. At present, five biosimilars have gained approval in both the European Union and the United States.4,5
This application describes the utilization of a nonreducing RPLC-MS workflow for intact mass comparison of trastuzumab innovator and biosimilar. The report includes the correlation of de-charged, observed masses with theoretical masses.
A recombinant human antibody standard, SILu™Lite SigmaMAb™ (MSQC4), is utilized as a reference and assay control sample. Here, this is referred to simply as SigmaMAb™ although several other SigmaMAb™ standards are also available commercially.
Workflow for Intact Mass Analysis of Trastuzumab
Antibody reduction (optional)
Chromatography
Measurement and Analysis
A complete RPLC-MS workflow has been developed to simplify intact mass analysis of non-reduced monoclonal antibodies (mAbs) for biosimilarity assessment.
In detail, it includes:
- Mass spectrometer calibration
- System suitability test utilizing a recombinant human monoclonal antibody reference
- A generic RPLC-MS method compatible with sample separation and analysis of both non-reduced and reduced monoclonal antibodies
Experimental - Sample and Reference Preparation and System Setup
Instrument Calibration
The Waters QToF Xevo G2XS mass spectrometer was calibrated in a mass range of 400 – 4000 m/z with a 10 μL/min infusion of 0.4 mg/mL of sodium iodide in water. Alternatively, calibration can be performed with a 10 μL/min infusion of 0.4 mg/mL cesium iodide, polyalanine in water prior to running the samples.
System Suitability
To evaluate performance of the entire workflow, an assay control (SigmaMAb™) was prepared and analyzed along with the samples. SigmaMAb™ reference was tested to ensure system suitability.
RPLC-MS System Setup and MS Data Analysis
RPLC-MS System Setup
The essential settings of the UHPLC-PDA chromatography system and the qToF mass spectrometer applied in the analysis of non-reduced antibodies are listed in Tables 1 and 2 below.
MS Data Analysis
Data were processed using the MaxEnt1 module within the UNIFI software to generate and analyze deconvoluted (zero charged) mass spectra. In general, a summed spectrum was created from the corresponding total ion chromatogram (TIC) of the eluted intact mAb. The summed m/z spectrum was then processed by the MaxEnt1 algorithm; detailed parameters are listed in Table 3.
For glycoform analysis, data were processed using UNIFI software from Waters™. Glycoforms were matched by the software. Glycoform relative abundance data were tabulated based on peak intensities of the coeluting glycoform species.
Results
The analysis objective here, was to perform non-reduced RPLC-MS intact mass analysis on all submitted samples to compare the molecular weight of trastuzumab innovator and biosimilar drugs.
Intact SigmaMAb™ was used to determine system suitability. All mAb samples were solubilized in 100 μL water to a final concentration of 1 mg/mL.
System Suitability Test Results
SigmaMAb reference sample (1 μL) was injected on the RPLC-MS system. Figure 1 illustrates the photodiode array (280 nm) and TIC (total ion current) traces of the non-reduced antibody, while Figure 2 displays the deconvoluted mass spectrum of the SigmaMAb™ reference. The observed intact mAb glycoforms matched the common glycoform masses of MSQC4, as listed in Table 4. The measured discrepancies between the observed masses and the theoretical values for four glycoforms are all within 0.011 % mass error.
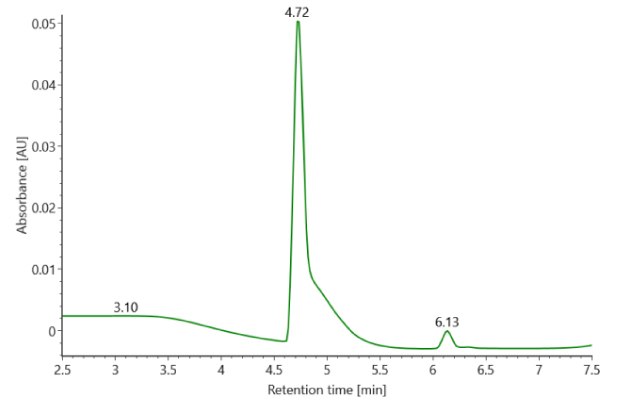
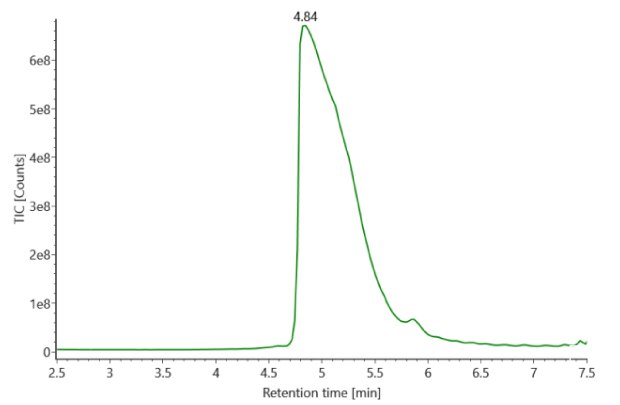
Figure 1. Photodiode array (280 nm, left) and TIC traces (right) of non-reduced SigmaMAb™ reference.
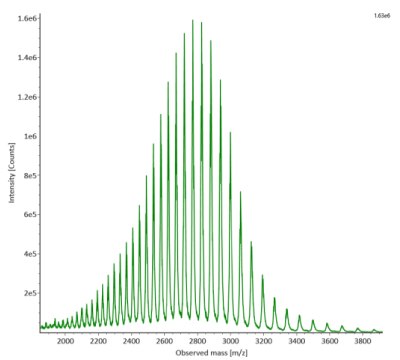
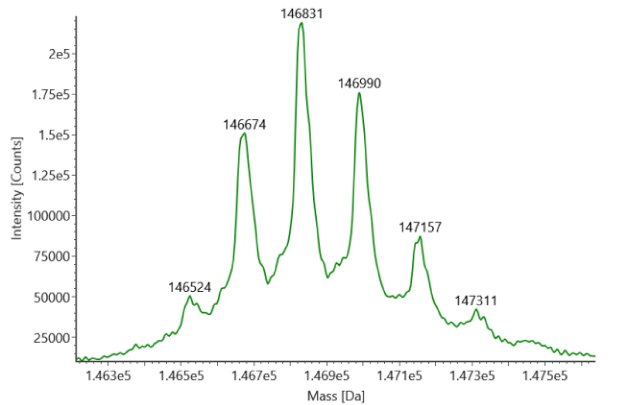
Figure 2. MS data for non-reduced SigmaMAb™ reference (MSQC4). Left: summed spectrum; right: deconvoluted spectrum.
Non-Reduced Sample Results
The monoclonal antibody samples were analyzed in their non-reduced form using RPLC-MS. The corresponding photodiode array (280 nm) traces, TICs, MS spectra and the deconvoluted MS spectra of trastuzumab innovator and biosimilar are shown in Figures 3 and 4. The relative quantitation results for N-glycoform distribution are shown in Figure 5. The method detects differences in the distribution of major N-glycoforms between innovator and biosimilar products. The observed masses of the non-reduced mAb correlate well with the calculated theoretical masses for all submitted samples, as shown in Table 5, and the observed mass error is 0.004% or less.
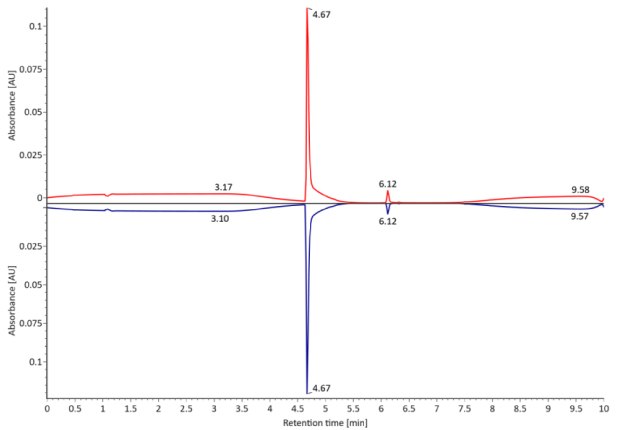
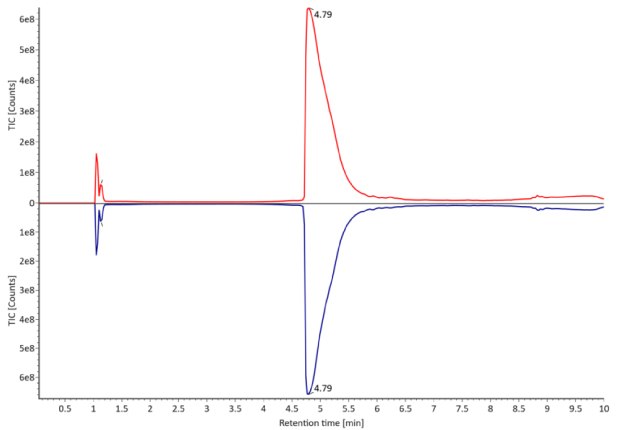
Figure 3. Comparative photodiode array (280 nm, left) and TIC traces (right) of non-reduced trastuzumab. Red: Innovator; blue: Biosimilar.
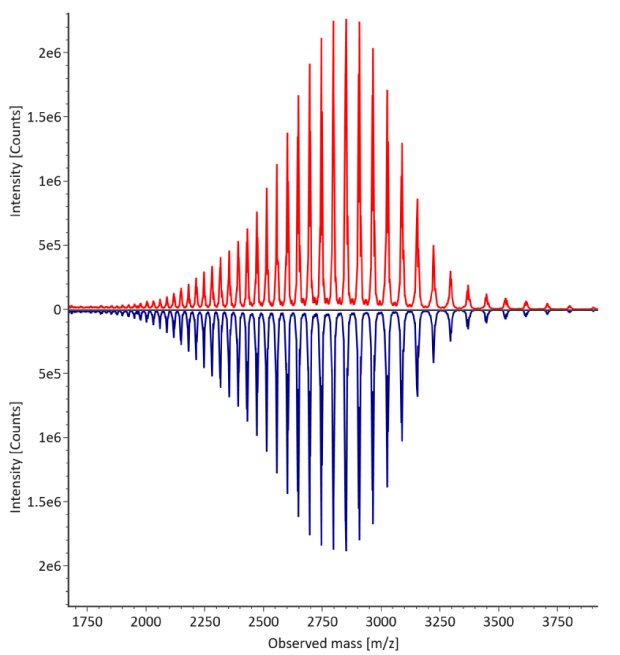

Figure 4. Comparative MS data for non-reduced trastuzumab. Left: summed spectrum; right: deconvoluted spectrum. Red: Innovator; blue: Biosimilar

Figure 5.N-glycoform distribution of trastuzumab by Intact RPLC-MS. Red: Innovator; Blue: Biosimilar.
Conclusion
Ensuring the regulatory approval of biosimilar monoclonal antibody (mAb) drug products necessitates a meticulous and effective comparability assessment to guarantee their safety and efficacy. This application note serves as an illustration of the utilization of reversed-phase liquid chromatography-mass spectrometry (RPLC-MS) at the intact level. The primary objective revolves around assessing the comparability between the original trastuzumab drug and its biosimilar counterpart. A comprehensive workflow was devised for intact mass analysis of non-reduced monoclonal antibodies, using trastuzumab as a representative mAb, and a SILu™Lite SigmaMAb™ product, as an assay control.
A separate workflow (Application Note: Biosimilar Comparability Workflow by Middle-Up Mass Analysis Using Trastuzumab as Model Protein - Protocol for reduction and RPLC-MS analysis of a monoclonal antibody) describes an optional mAb reduction procedure which can be performed with either a partial, or full, reduction procedure. Depending on the procedure used, intra-chain disulfide bonds remain intact (partial reduction) or are broken (full reduction). Solutions of sodium iodide, or cesium iodide, were used for mass spectrometer calibration, and a system suitability test, utilizing a recombinant human monoclonal antibody reference, were developed for routine use. In addition, an RP method suitable for separation and analysis of both reduced and non-reduced mAbs was established using a wide-pore, superficially porous C4 column.
Results obtained from the non-reduced SigmaMAb™ reference indicated minimal disparities of 0.011% mass error, or less, between observed and theoretical masses for four glycoforms. For trastuzumab, both the innovator and biosimilar samples displayed a strong concurrence between observed and theoretical non-reduced mAb masses, with an observed mass error of 0.004%, or lower.
The experimental findings affirm the versatility of the workflow for analyzing non-reduced monoclonal antibody samples, delivering accurate results that enable distinct identification of multiple glycoforms.
See more applications on the Biopharmaceutical Characterization page.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?