Antibody-Enzyme Mediated Hot Start PCR Protocol
PCR Technologies Protocols Table of Contents
End-point PCR Protocols
During PCR assay preparation, nonspecific amplification can occur due to binding of PCR primers to nonspecific templates and from formation of primer dimers which result from using other primer molecules as templates. The protocols presented in this section both adopt controlled PCR activation to allow users to minimize nonspecific amplification while increasing target yield and specificity. Two alternative methods are presented to control amplification of the specific product, these are enzyme and dNTP mediated Hot Start PCR.
When using hot start Taq DNA polymerase, the enzyme remains inactive until heated. Hot Start DNA polymerase control is achieved by chemical or antibody modification of the enzyme. Chemically modified hot start enzymes require up to 10 minutes activation whereas antibody mediated hot start enzymes are activated within 1 minute.
JumpStart™ Taq DNA Polymerase is an antibody-inactivated, hot start enzyme. During the initial denaturation step of the PCR, the antibody is also denatured and dissociates from the DNA Polymerase, therefore enzyme activity is restored. The resulting PCR exhibits a higher specificity and yield5.
Equipment
- Pipettes dispensing volumes from <1 to 200 μL
- Benchtop microcentrifuge
- Thermal cycler
Appropriate Analysis Equipment
- Electrophoresis equipment
- UV transilluminator
- Alternative PCR product analysis system
Supplies
- Sterile filter pipette tips
- Sterile 1.5 mL screw-top microcentrifuge tubes (CLS430909)
- PCR tubes and plates:
• Individual thin-walled 200 μL PCR tubes (Z374873 or P3114)
• Strip tubes, 200 μL (Z374962)
• 96-well plates (Z374903)
• 384-well plates (Z374911) - dNTP mix, 10 mM each of dATP, dCTP, dGTP, and dTTP (D7295).
- Enzyme and buffer; review Table P2-1 to select the optimal reagents for the experiment.
- If using gel electrophoresis for PCR product analysis, a DNA size marker is usually required. Select the appropriate marker based upon PCR amplicon size (Figure 2‑17).
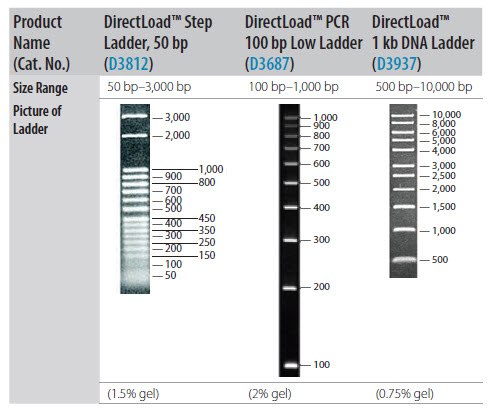
Figure P2-17.Resolution of DNA Size Standards Through Agarose Gel.
- PCR grade water (W1754 or dispense W4502 into 20 mL aliquots and freeze; use a fresh aliquot for each reaction).
- DNA/cDNA template:
- cDNA reaction diluted 1:10 to detect medium to highly expressed targets or between 1:2 to 1:5 for rare transcripts.
- gDNA 10 ng to 100 ng.
- Primers diluted to working concentration (10 μM working stocks are appropriate for most assays but multiplex blends may require mixing of 100 μM stocks).
- Custom oligos can be designed according to the OligoArchitect Online (OligoArchitect Assay Design) and can be ordered here.
Method
- Leaving the DNA polymerase on ice or at –20 °C, thaw the remaining reaction components on ice, vortex to mix (except the enzyme), centrifuge briefly and replace on ice.
- Set up PCR reactions:
- Prepare a master mix containing all reaction components with the exception of the DNA/cDNA template (Tables P2‑2A or P2‑2B). Calculate the master mix required by multiplying amounts by the number of reactions needed, including controls and then add 10% to ensure a sufficient quantity for all samples.
*Note: Magnesium chloride is added separately if not already in the PCR buffer or when previous optimization has revealed a requirement for a higher concentration.
b. Combine reaction components into a 1.5 mL microcentrifuge tube on ice.
- Mix the reaction master mix by carefully pipetting up and down, ensuring that all mix is expelled from the pipette tip and then pulse or centrifuge briefly to collect the sample at the bottom of the tube.
- Aliquot 20 μL of master mix into the required numer of 200 μL thin-walled PCR tubes (label/number the tubes containing samples, including replicates and controls).
- Add 5 μL of the DNA template sample (containing a total of 10 ng to 100 ng gDNA or dilute a cDNA sample 1:2 to 1:10) to reach a final reaction volume of 25 μL.
- Spin the PCR tubes and place into a thermal cycler with a heated lid.
- Determine the appropriate Ta for the primers. A good first test can be performed using a Ta that is 5 °C lower than the Tm of the primer with the lowest Tm.
- Determine the required thermal cycling protocol with reference to Table P2-3.
- End the PCR with an incubation at 72 °C for 10 min to ensure that all products are full length.
*Note: Elongation time is dependent upon amplicon size: 30 sec for up to 500 bp. Add 1 min for each additional 1 kb.
- Analyze a 10 μL aliquot of the completed reaction by agarose gel electrophoresis, with visualization on transilluminator or other chosen analysis method.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?