Peptidoglycan Structure, Biosynthesis and Function
Peptidoglycan Structure
The basic structure of peptidoglycan (PGN) contains a carbohydrate backbone of alternating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid, with the N-acetylmuramic acid residues cross-linked to peptides. Peptidoglycan provides rigidity to the cell wall; the cell walls of Gram-positive bacteria may contain up to 40 layers of peptidoglycan, conferring significant mechanical strength.1
Structurally, bacteria resemble primitive plants in that the cellular contents are surrounded by an inner peptidoglycan cell wall in addition to an inner plasma membrane and, in Gram-negative bacteria, an outer lipid bilayer (Figure 1). In Gram-negative bacteria, peptidoglycans make up about 10% of the cell wall dry weight; while in Gram-positive bacteria the thicker peptidoglycan layer contains about 20% of the cell wall dry weight. Although the peptidoglycan is responsible for the mechanical strength and shape of bacterial cells, it has sufficient plasticity and dynamic turnover to allow cell growth and division.
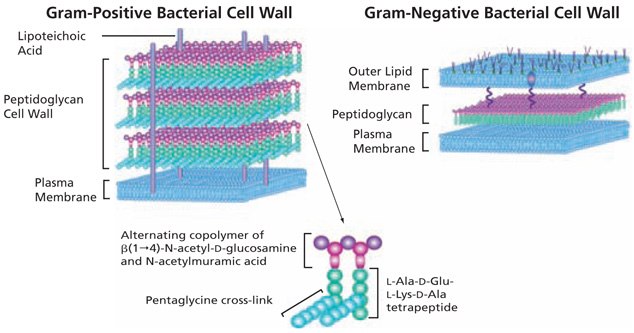
Figure 1.The basic structure of bacterial peptidoglycan and the cell wall structures of Gram-positive and Gram-negative bacteria.
Peptidoglycan Biosynthesis and Degradation
The peptidoglycan biosynthetic pathway begins in the cytoplasm with the synthesis of a muramyl peptapeptide precursor containing a terminal D-Ala-D-Ala. L-Alanine is converted to D-alanine by racemase, with subsequent assembly of D-alanyl-D-alanine by D-Ala-D-Ala ligase. In the cytoplasm, the muramyl pentapeptide precursor is anchored via a water-soluble UDP-glucosamine moiety. In the second phase of peptidoglycan construction, the muramyl pentapeptide N-acetylglucosamine is transferred to a C55 undecaprenyl phosphate with the release of UMP to form a Lipid I intermediate. An additional glycosylation step completes the peptidoglycan unit, which is then transported via its C55 lipid tail to the external periplasmic surface of the membrane, where the peptidyglycan unit becomes integrated into the cell wall matrix. Several transpeptidases and transglycosylases connect the newly formed peptidoglycan structures to the cell wall peptidoglycan matrix.
Inhibitors of peptidoglycan biosynthesis act as antibiotics in that they lead sequentially to the loss of peptidoglycan, loss of cell wall integrity, and lysis. Peptidoglycan degradation is catalyzed by glycosidases, peptidases, and amidases, including lysing enzymes such as lysozyme, that are commonly used for degrading cell walls.
Specific antibacterial compounds that contain a β-lactam structure, such as penicillin, interfere with the synthesis of the cell wall, weakening the peptidoglycan scaffold within the bacterial wall so the structural integrity eventually fails. Since mammalian cells have a plasma membrane but lack the peptidoglycan wall structure, this class of antibacterials selectively targets bacteria with no significant negative effect on the cells of the mammalian host. The specificity of β-lactam antibacterials is due to their structural similarity to the D-alanyl-D-alanine group, allowing them to compete for the binding sites of transpeptidases and prevent the assembly of peptidoglycan layers in both Gram-positive and Gram-negative bacteria.
Peptidoglycan Functions
The primary immune recognition is based on structures common among invading pathogens. Surface molecules, such as lipopolysaccharide (LPS), peptidoglycans, and peptidoglycan recognition protein (PGRP), are known to elicit immune reactions ranging from cytokine release to fever.2‑4 Peptidoglycans activate the Toll-like receptor 2 (TLR2), that is present in mammalian cells, and they can be used for the stimulation of lymphocytes. Peptidoglycans also function as antagonists of Poly(I:C).5 Peptidoglycans may be used to estimate the activity of lysing enzymes such as lyticase.6
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?