FLAG® HA System
Isolation and analysis of protein complexes continues to be a major technical challenge in function proteomics research. Co-immunoprecipitation is the most widely used method for isolation of protein complexes. The TAP (tandem affinity purification) methodology incorporates tandem-linked affinity tags into a target protein to pull down and isolate endogenous interacting proteins. Protein complexes isolated using TAP have a much higher purity compared to single purification.
Overview for FLAG® HA System
The FLAG® HA tandem epitope system consists of small epitopes tags that are not eukaryotic derived. These features minimize interference with protein functions and provide superior specificity. The hydrophilic character of the FLAG® HA peptide increases the likelihood that the dual epitope will be located on the surface of the fusion protein where it is accessible for the antibody-antigen interaction.
Other TAP systems, including combinations of GST, clamodulin, nickel binding proteins, streptavidin, and Protein A have suffered from several limitations, such as interference with complex assembly or protein function due to relatively large TAP tags (20 KD and larger); high rate of contamination with non-targeted endogenous proteins; and requirement of TEV protease treatment for elution which adds additional bacterial contaminants.
Key Features of the FLAG® HA System
- Affinity resins have high specificity and pull down less contaminates compared to existing systems
- The TAP tag does not sterically affect the target proteins’ interactions
- No need for protease elution
- Efficient and mild elution for the first immunoprecipitation and flexible elution strategies for the second
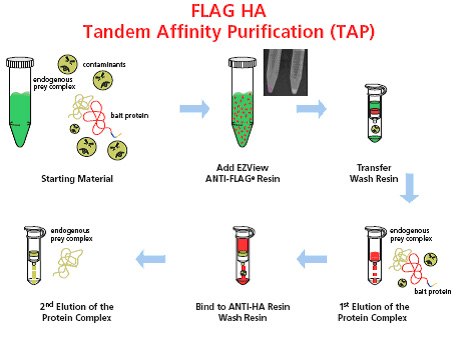
Figure 1. Schematic representation of the FLAG® HA Tandem Affinity Purification system. The whole procedure involves six simple steps: 1) Prepare the starting material that contains FLAG® HA tagged bait protein; 2) Add EZView ANTI-FLAG® resin directly to lysate; 3) Transfer resin to a spin column and wash; 4) Conduct first elution with 3X FLAG® peptide; 5) Transfer the first eluate directly to a spin column containing anti-HA affinity resin, bind and wash; and 6) Conduct second elution with elution buffer depending on downstream manipulations (urea, HA peptide, or Laemmli Sample Buffer).

Figure 2. Specificity analysis of FLAG® and HA tags and affinity resins using plant materials. Panel A shows high specificity of the ANTI-FLAG® and anti-HA antibodies to FLAG®- and HA-tagged fusion proteins, respectively, in 7 different plant extracts (tabacco, Arabidopsis, maize, soybean, rice, tomato, and cotton). Immunoblot detection was with ANTI-FLAG M2®-AP and anti-HA-HRP antibody conjugates. Panel B shows specificity of ANTI-FLAG® and anti-HA resins in comparison with existing resins (ANTI-FLAG®, anti-HA, Protein G, CBP, Streptavidin, Nickel, and Glutathione). After washing, proteins that were non-specifically bound to the resins were eluted, resolved on PAGE gel, and detected by silver stain. Both ANTI-FLAG® and anti-HA resin showed minimal cross-reactivity compared to other resins. Panel C illustrates consecutive immunoprcipitations provide a high purity of sample compared to a single immunoprecipitation.
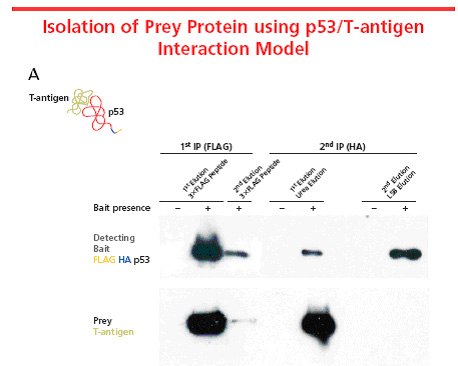
Figure 3. A well-known protein-protein interaction system in mammalian cells, p53 and T-antigen, was used to characterize the FLAG® HA system. FLAG® HA tandem tag was incorporated into p53 using FLAG® HA Tandem TAP Tag Generation Kit (TP0020) and transiently expressed in COS-7 mammalian cells. The tagged p53, used as the bait protein, interacts with the endogenous T-antigen to form a complex. The protein complex was isolated using the TAP procedure. The figure illustrates that FLAG® HA does not interfere with the interaction. In this case, preferential elution of the T-antigen was performed using a urea-based eluatant. Elution with the 3X FLAG® peptide eluted both proteins from the resin equally. Elution conditions vary based on proteins and expression systems being used. Optimization of elution conditions is required for different applications and downstream analyses.
Related FLAG®-HA Products
To continue reading please sign in or create an account.
Don't Have An Account?