Protein Binding Determination Using Supel™ BioSPME C18 Pin Device as Part of the Manual Methodology With and Without Shaking
Introduction
A comparison of protein binding results from two different sample preparation procedures was carried out. The extraction step was performed with and without agitation and the results were compared with regard to the accuracy of protein binding values and extraction reproducibility. The method employed was based on using a manual operation procedure.
Experimental
Experimental data involving two variations for the sample extraction step will be discussed since all other sample preparation steps will be held constant.
Definitions:
- Static – no movement of coated pins and well plate while coated pins are immersed during processing
- Shake – refers to the orbital shaking movement of the coated pins and/or the well plate during the extraction step only. The shaking parameters were orbital radius (1mm) and orbital speed (1250 rpm)
A generic method for using the BioSPME C18 pin device with 200 μL sample volume is outlined below. By clicking on the step descriptions the conditions used for this study to determine the free/bound fraction of drug in human plasma study are displayed. The extraction is performed from both buffer and plasma samples and protein binding is calculated using the determined extracted amounts.
Steps for BioSPME Determination
Condition
20 minutes in 100% 2-propanol (400 µL)
1st Wash
10 seconds in 100% water (500 µL)
Extraction
15 minutes (static or shaking@1250 rpm) (200uL)
2nd Wash
1 minute in 100% water (500 µL)
Desorption
15 minutes in 80:20 (v/v) methanol:water (200 µL)
Analyze by LC-MS/MS
Analyze by LC-MS/MS
Analyte concentration levels used corresponded to the therapeutic drug levels, each analyte was spiked and analyzed separately:
- Carbamazepine: 100 ng/mL
- Diazepam: 100 ng/mL
- Prednisolone: 500 ng/mL
- Zolpidem: 100 ng/mL
Basic LC-MS/MS analysis with Biphenyl HPLC Column
Prepare five instrument calibration standards in 80:20 methanol:water to cover an expected range of analyte concentrations in the final extract. Internal standard carbamazepine-D10 was used in the desorption solution. These were injected along with the extracted analytes and used to determine the mass extracted (Table 1 and 2).
The percent protein binding for each studied analyte was used to evaluate the effectiveness of the extraction step variation. The percent free or percent unbound is determined in Equation 1.

Concentration free represents the unbound concentration of the analyte in the matrix in this case plasma, and concentration total represents the total concentration of analyte. The amount extracted is independent of units and can be applied using preferred quantities (e.g. nanograms or moles) Mfree, and extraction volume of plasma, Vplasma. The concentration of analyte in the desorption solution is quantified by an external calibration curve, and if the desorption volume is equal to the plasma and buffer extraction volumes, the concentration from desorption will be equal to the extracted concentration as shown in Equation 2.

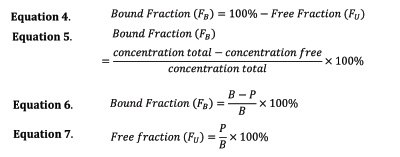
The bound fraction, FB, can be determined from the extracted concentrations as shown in Equation 6.
In cases where depletion of compounds from plasma is pronounced upon BioSPME extraction (extraction exceeded 5% of total spiked analyte), a correction to the calculated Bound Fraction is required as described below:

B and P, represent the respective amounts extracted from buffer, B, and plasma, P. P0 and B0 represents the concentration the samples were spiked or the total concentration in plasma or buffer. Equation 8 accounts for the concentration in solution after extraction on the fiber: the depletion of the analyte from Sample. Equation 6 and Equation 7, do not take this consideration into account. However, they provide accurate values when the extracted amount from plasma is less than 5% of the total spiked amount.
Results
The experimental sample size (n) was set at 8. The tables below show extracted amounts and protein binding values for both buffer and plasma sample sets. Relative standard deviation (RSD) for each extraction is shown.
Better accuracy for protein binding values was achieved when using shaking during extraction as evident in Figure 1 where the experimentally determined values are compared to these from literature.
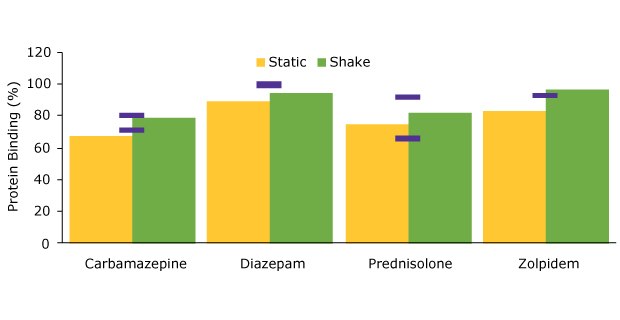
Figure 1.Comparison of protein binding values for static vs. shake manual extraction.
The added benefit of better precision is shown when comparing the RSD values for both the buffer and plasma extractions as summarized in Figure 2.
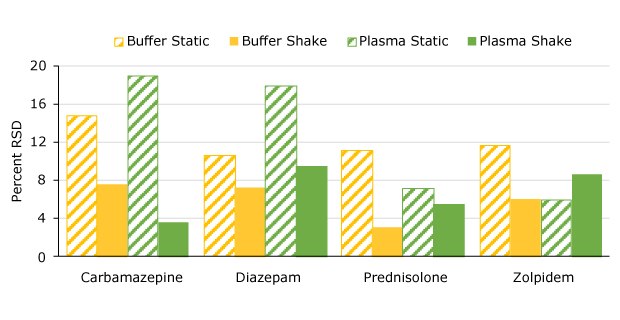
Figure 2.Precision comparison (%RSD) static vs. shake manual extraction.
Conclusion
In processing of both buffer and plasma samples, evidence indicates that the extraction step performance is improved when shaking is used and directly compared to static conditions. The use of static extraction step in the manual methodology yielded no workflow advantage in terms of time and simplicity. Lower amounts extracted using static conditions in comparison to agitation (shake) indicate that uptake to equilibrium is not achieved under static conditions using the selected extraction time. Exposure of the pin tool to more sample matrix using shaking helps in increasing the mass transfer rate to achieve faster equilibrium. Addition of an appropriate heater/shaker to the method results in better accuracy and precision of results.
To continue reading please sign in or create an account.
Don't Have An Account?