Reverse Transcription
One approach to the analysis of gene expression is to measure the concentration of mRNA of a gene. There are several challenges to such analysis, such as the differences in half-life between different transcripts, the temporal patterns of transcription and the lack of correlation between mRNA and protein. To analyze RNA using a PCR-based method, cDNA must be produced using reverse transcription (RT). This process utilizes a reverse transcriptase enzyme and dNTPs.
The RT step may be performed on total RNA, such that a global cDNA representation of many transcripts is produced (usually via a two-step protocol) or in a gene-specific approach in which only the RNA of interest is converted to cDNA (usually following a one-step protocol).
Since it has been demonstrated that the two-step RT reaction is not always linear with respect to input RNA and cDNA yield1, it is important to determine and control the total amount of RNA extracted and included in RT reactions. Measuring the concentration of RNA presents uncertainty and the absolute value is dependent on the instrument or system used to make the measurement. As can be seen in Figure 8.1, there is a large variability between concentration measurements of 5 samples of RNA (A–E) (see Sample Purification and Quality Assessment) when using the Nanodrop, conventional UV spectrophotometry, Ribogreen, the Agilent 2100 BioAnalyzer, or the Bio-Rad Experion. Note that the presence of EDTA (samples D and E), which would inhibit downstream RT and PCR, and degradation (sample C) have different effects on concentration measurements depending on which system is used. This observation is illustrative of the importance of performing additional quality control steps prior to using samples in downstream reactions (see Sample Purification and Quality Assessment).
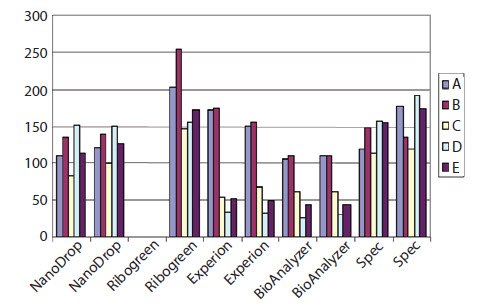
Figure 8.1.The concentration of five samples of total RNA (A–E) was measured using Nanodrop, conventional UV spectrophotometry, Bio-Rad Experion, the Agilent 2100 BioAnalyzer (all duplicate measurements) or Ribogreen (single measurement). As can be seen, the absolute concentration as well as the relative concentration varied between the samples. This was because sample C was degraded and samples D and E contained EDTA, which caused inaccurate measurements in the Experion and BioAnalyzer systems.
Reverse Transcription Linearity
The relative concentration of total RNA can influence the efficiency of the RT and the concentration of cDNA produced from a given transcript. Therefore, it is desirable to include the same or a very similar concentration of RNA into all two-step cDNA synthesis reactions, unless the RT system has been verified to have a linear response. As can be seen in Figure 8.2, using a conventional RT protocol, the 100-fold dilutions of input RNA do not result in a corresponding 100-fold difference in cDNA yield for the templates tested. Interestingly, the data presented are duplicate qPCR run on duplicate RT reactions. As shown, the lack of linearity is reproducible between the two RT reactions.
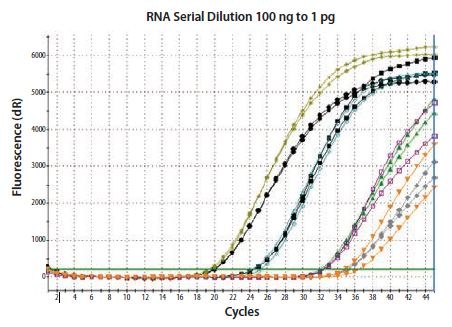
Figure 8.2.Total RNA was diluted through 100-fold and reverse transcribed using two-step random priming; two independent RT reactions were performed. β-actin was detected in duplicate qPCRs for each RT reaction. The RT is reproducible, but cDNA yield is not proportional to input RNA concentration. Therefore, if the experimental restraints require that a variable concentration of RNA is included in the RT, it is critical to verify that the protocol and reagent combination result in a linear response.
In the example shown in Figure 8.3, ReadyScript® RT reagent (RDRT) was used to reverse transcribe total RNA from a 2-fold and a 10-fold serial dilution of template using a two-step protocol and a combination of oligo-dT (O4387) and random priming (described below). The CANX gene was detected in both dilution series with a direct proportionality to the input RNA concentration.
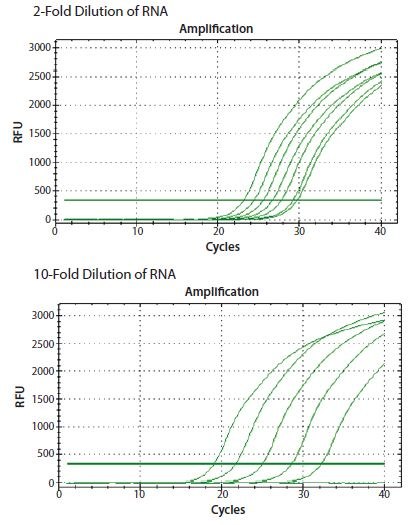
Figure 8.3.ReadyScript® RT reagent (RDRT) was used to reverse transcribe total RNA from a 2-fold and a 10-fold serial dilution. The gene CANX was detected in both dilution series resulting in a direct proportionality to the input RNA concentration (data from student groups attending an EMBL Advanced qPCR Workshop).
Reverse Transcription Priming
The choice of primers to be used to initiate reverse transcription can greatly affect RT-qPCR results. For one-step
RT-qPCR, gene-specific primers are used. When performing a two-step assay, a reverse gene-specific primer, oligo-dT (O4387), random hexamers, nonamers, decamers, dodecamers or pentadecamer2 or a combination of oligo-dT (O4387) and random primers may be used. Gene-specific priming is usually run in separate reactions for each target RNA. These separate reactions may have very different efficiencies, thus complicating comparisons between RNA concentrations. On the other hand, when using gene-specific primers, all of the RT product will encode the gene of interest and may allow quantification of very low abundance mRNAs that could not be detected using nonspecific RT primers. To avoid the potentially high inter-assay variations in RT that can occur with gene-specific primers, nonspecific primers may be used to generate a pool of cDNA. Separate qPCR assays for each target can be performed subsequently with aliquots from the cDNA pool. If all qPCR targets are near the 3’-end of polyadenylated mRNAs, oligo-dT (O4387) is a suitable primer choice. On the other hand, if the qPCR targets are more than a few kilobases from the 3’-end or if the RNA is not polyadenylated, random primers will result in more reliable detection. If the relative 3’ location of qPCR targets varies or the desired transcripts contain a combination of polyadenylated and non-polyadenylated RNAs, a mixture of oligo-dT (O4387) and random oligomers will give the best results
Reverse Transcription Priming for Two‑step RT Reactions
Two approaches are commonly utilized for two-step RT priming; oligo-dT (O4387) and random priming. The oligo-dT (O4387) method relies on hybridization of an oligodT (usually 15mer) to the poly-A tail, present on the 3’ end of the majority of mRNA molecules, to prime and selectively reverse transcribe mRNA.
This approach, while conceptually very simple, has associated challenges: The oligo-dT (O4387) primer binding is not specific to mRNA at the reaction temperatures used for the RT, thus the oligo-dT (O4387) will bind nonspecifically to other regions of RNA. Additionally, stretches of rRNAs are also detected because ATrich regions in these molecules are primed by oligo-dT (O4387). Some mRNAs, like those encoding histones, do not contain poly A tails and might not be represented within the resultant cDNA.
The second method uses random priming. Random primers consist of random sequences, frequently of hexamers (6mer) or nonamers (9mer). These are used to prime the RT reaction, leading to the synthesis of cDNA fragments of varying length, which represent the original RNA. Random primers hybridize along the length of the transcript and tend to be more tolerant of secondary structure than oligo-dT (O4387) or gene-specific priming.
To benefit from the advantages of the respective techniques, some protocols call for a combination of both primer types. A specific primer to the target sequence may also be used in a two-step RT protocol but is more often used in a one-step procedure (see below).
Reverse Transcription Priming for One‑step RT Reactions
In a one-step RT protocol, gene-specific primers are used to reverse transcribe a single target. The design of the genespecific primer is critical; it must lie within an open, accessible region of the mRNA target when predicted at the temperature of the RT reaction. Under these conditions there is a linear relationship between input RNA and cDNA (Figure 8.4). This primer may be (and usually is) common to the PCR primer.
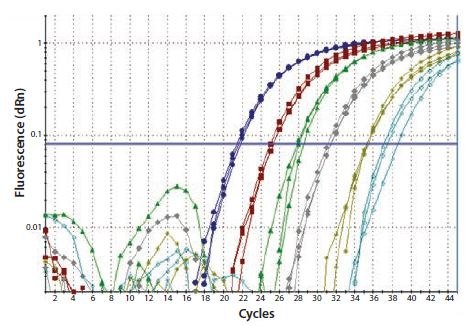
Figure 8.4.Total RNA was diluted through a serial 10-fold dilution series and a one-step RT-qPCR performed to detect GAPDH. Each reaction was performed in triplicate. This resulted in a linear relationship between the RNA concentration and the cDNA yield (image courtesy of Prof. Stephen Bustin, Anglia Ruskin University, UK).
Reverse transcription is a highly variable process and all steps must be considered to keep reaction components as constant as possible. Performing a one-step RT-qPCR reaction reduces the number of pipetting steps required and will reduce error. It may, therefore, be the method of choice if small differences need to be measured and accuracy is paramount. However, the contrary consideration is that the determination of the gene of interest (GOI) to reference gene ratio (see Data Analysis) requires two separate one-step RT reactions rather than a single cDNA from a two-step reaction and the two targets cannot be detected using a multiplex qPCR approach.
Reverse Transcription Efficiency
It is generally assumed that all the RNA/mRNA in a RT reaction is converted to cDNA and that all transcripts are converted in a 1:1 ratio or proportionally to the starting RNA concentration. Recent studies have been performed to investigate each of these assumptions. It is clear that that the amount of RNA that is converted into cDNA is highly variable. The two-step RT process is variable and specifically dependent on RNA concentration, enzyme, buffer composition and priming protocol. Since the process is variable, it is important to maintain as many constant conditions as possible1,3,5.
For two-step RT reactions, in general it is necessary to aim for the same input RNA concentration as well as to keep the priming conditions, RT enzyme and buffer constant. When a constant input concentration cannot be determined, it is advisable to use a one-step process, include a carrier such as Polyethylene Glycol (PEG)6, or select a commercial kit that has been validated to result in a linear response, such as ReadyScript® RT.
The choice of priming strategy influences both the absolute yield and ratio of different cDNA targets from a total RNA sample. Figure 8.5 shows the cDNA copy number yield for three different genes (shown by the color of the histogram) in a RNA sample that has also been subjected to controlled, enzymatic degradation. It is clear that the different priming methods result in different absolute copy numbers and ratios between the samples. In addition gene specific priming appears to be more tolerant of RNA degradation than oligo‑dT priming.
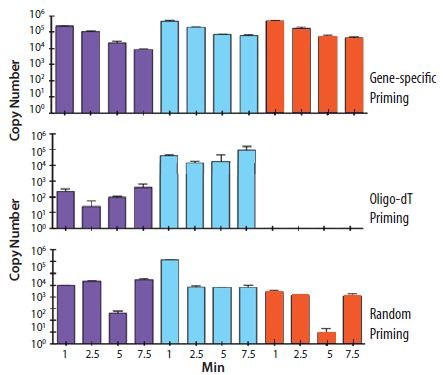
Figure 8.5.Total RNA was incubated on a naked human hand for the times indicated (1, 2.5, 5 and 7.5 min). cDNA was prepared using gene specific, random or oligo-dT (O4387) priming. The copy number of three genes was determined, as indicated by the purple, turquoise and orange histograms. It is clear that the different priming strategies influence the detection of each gene with oligo-dT (O4387) priming resulting in no detection of gene 3 (orange) which is clearly apparent in the RNA sample since it is detected using gene specific priming (data provided by Prof. Stephen Bustin, Anglia Ruskin University, UK).
The temperature used for RT reactions may affect specificity, especially when hybridizing gene-specific primers. Primers that can form a strong 3’-duplex will hybridize more readily at lower temperatures. Since RT enzymes can extend from a DNA primer on a DNA template, primer-dimer formation may start during the RT step. Increasing RT incubation temperature to the highest temperature at which the enzyme is fully active or using a high-temperature enzyme may reduce the amount of primer-dimer product. The primers used in Figure 8.6 resulted in significantly more specific product relative to nonspecific product after one-step RT-qPCR when RT was performed with Moloney Murine Leukemia Virus-Reverse Transcriptase (MMLV-RT) at 45 °C (Figure 8.6B) than they did when RT was performed at 37 °C (Figure 8.6A). Similarly, performing twostep RT-qPCR with a nonspecific primer for RT and Hot Start Taq polymerase for qPCR resulted in less primer-dimer product (Figure 8.6D) than using one-step RT-qPCR with gene specific primers that could form a 3’-duplex (Figure 8.6B).
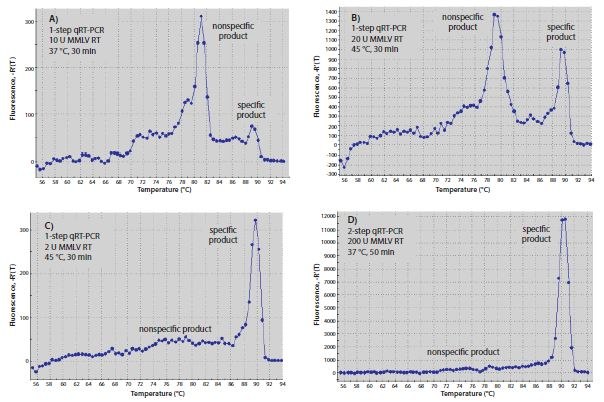
Figure 8.6.Optimization of RT. (A–C) Melt curves of RT-qPCR products produced with one-step or (D) two-step RT-qPCR. Reactions A–C each contained 10 μL of SYBR® Green JumpStart™ Taq ReadyMix™, 0.02 μL of Reference Dye, both gene-specific primers at 0.4 μM, and 10 ng human total RNA in a final volume of 20 μL. Genespecific primers were 5’-CGGGCTTCAACGCAGACTA-3´and 5´ -CTGGTCGAGATGGCAGTGA-3´ for c-fos (Accession NM_005252). Reactions A and B also contained 20 units of MMLV- RT, whereas reaction C contained 2 units. Reaction A was incubated at 37 °C for 30 min before qPCR, whereas B and C were incubated at 45 °C for 30 min before qPCR. In D, the RT reaction contained 1x MMLV buffer (Product No. D8559), 0.5 mM dNTPs, 1 μM oligo-dT (O4387), 0.8 units/μL RNase inhibitor, 200 units MMLV-RT, and 10 ng human total RNA in a final volume of 20 μL. The reaction was incubated at 25 °C for 10 min, 37 °C for 50 min, and 80 °C for 10 min. 2 μL of the RT reaction product were added to qPCR containing 10 μL of SYBR® Green JumpStart™ Taq ReadyMix™ 0.02 μL of Reference Dye, and both gene-specific primers at 0.4 μM as for the one-step reactions (A–C). All qPCR reactions were incubated at 94 °C for 3 min to denature, then for 40 cycles of 94 °C for 15 sec and 60 °C for 1 min.
The amount of RT enzyme per reaction can also affect RT-qPCR results. As shown in Figure 8.6, one-step reactions with 2 units of MMLV-RT (Figure 8.6C) were more specific than reactions with 20 units (Figure 8.6B). Two-step RT-PCR using oligo-dT (O4387) or random primers for RT, resulted in greater specificity than one-step RT-PCR (Figure 8.6D). This could be attributed to the fact that the gene-specific primers are not present during the low temperature RT reaction, thus preventing formation of nonspecific products. Higher concentrations of RT may give better results in two-step reactions, but because the RT enzyme can interfere with Taq DNA polymerase activity7, the amount of RT product transferred to qPCR should be limited to no more than 10% of the final reaction volume. The exception to this recommendation would be when using ReadyScript, in which 25% of the PCR volume can be RT reaction without affecting the PCR efficiency.
This description of the variables that are inherent in the RT process demonstrates that the determination of gene
expression from RT is dependent on the RT method used, the quantity and quality of the samples, in addition to the consideration of template quantity and should be carefully reported, as described in the MIQE guidelines8 (see Quantitative PCR).
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?