Fast Analysis of Monoclonal Antibodies Using Silica Monoliths Designed for Bioanalysis
Introduction
In the last two decades, the pharmaceutical market has changed dramatically from small molecules to protein-based drugs and antibodies, which have a higher potential for targeting extraneous substances.1 This is supported by the fact that six of the top ten best-selling pharmaceuticals are based on monoclonal antibodies (mAbs). In comparison to chemical entities, the analysis of mAbs is complex and requires several separation techniques, e.g., affinity chromatography, reversed phase chromatography, size exclusion chromatography and ion-exchange chromatography. In particular, the strong growth in development of biosimilars due to expiring patents of those blockbusters, led to increased requirements concerning column performance in HPLC.2
The analysis of biomolecules using conventional HPLC columns is usually associated with limited accessibility of surface for larger molecules, slow diffusion leading to extremely broad peaks with severe tailing, and possible conformational changes during elution. One of the reasons for those observations is the mesopore size ranging from 8-15 nm (80-150 Å). The separation quality could be improved by using column materials with wider mesopores of ca. 30 nm (300 Å) or larger.3-5
During product development, process monitoring, and quality control testing of mAbs, fast and precise analytical separations are necessary. The most important technique for determination of mAb titer is affinity chromatography using protein A. Protein A, a cell wall protein from Staphylococcus aureus, was the first isolated protein with a high affinity to the Fc region of immunoglobulin G (IgG).6 Its high selectivity and high resistance to temperature, pH and tryptic cleavage make protein A a powerful tool in antibody purification.7 Today, the use of protein A as a ligand on chromatographic media is one the most commonly employed methods for antibody capture during the purification process. In comparison to process protein A media, only a few vendors provide analytical protein A columns for process monitoring and quality control. Most of these columns are packed with particles limited in column backpressure, chromatographic performance, and application for matrix rich samples, such as harvested cell culture fluids. As a consequence, sample preparation is more complex and the limited column backpressure results in long analysis time due to lower flow rates.
Silica monoliths, which consist of one continuous piece of silica, are an alternative to fully porous or superficially porous particles, and are preferred for the application and fast separation of matrix-rich (“dirty”) samples due to their bimodal pore structure consisting of large through pores (macropores) and mesopores.8, 9 They are prepared according to a sol-gel process leading to a bimodal pore structure where both macropore and mesopore formation can be controlled individually.9, 10 The design of larger macropores offers good flow characteristics resulting in much lower column backpressure as compared to columns packed with particles, and maintaining column performance even at higher flow rates.11-13 Due to the hydrodynamic volume of mAbs (mostly 5-6 nm), the alteration of mesopore development up to a minimum of 30 nm (300 Å) is necessary to allow them to enter the pores for improved chromatographic separation and to prevent size exclusion effects. Recently, the development of silica monoliths containing larger mesopores, and their application for the analysis of biomolecules, has been reported,14 leading to the now commercially available Chromolith® WP 300 column family.
Another sector in chromatography getting more and more interest is the coupling of desired ligands for specific interactions with certain analytes, e.g., antibodies, aptamers or enzymes.15-20 The majority of described protein immobilizations were performed on polymeric particles or monoliths and silica particles. In the last decade, silica-based monoliths were also used for the immobilization of several ligands to enable their use in the applications of affinity chromatography, for chiral separations, or as on-column bioreactors.21-26 The development of wide pore silica monoliths enhanced the column properties for the immobilization and analysis of larger molecules. In this study, recombinant staphylococcal protein A (rSPA) was immobilized onto a silica monolith containing larger mesopores suitable for the separation of mAbs. The analysis performed on the monolithic column was characterized by different parameters including detector response linearity, reproducibility and long-term stability.
Experimental
Chemicals
See the reagents & chemicals listing at the end of this article. In addition, native recombinant staphylococcal protein A (rSPA) ligand was purchased from Repligen (Waltham, MA, USA), pure cetuximab stock solution was a research sample from Merck KGaA, Darmstadt, Germany, and gammanorm IgG was obtained from Octapharma (Heidelberg, Germany).
Epoxy-modified wide pore silica monolith (25 x 4.6 mm) columns were prepared as research samples at Merck KGaA, Darmstadt, Germany (now available as Chromolith® WP 300 Epoxy).
Apparatus
All modification and cleaning steps necessary for the immobilization of rSPA ligand on epoxy-modified columns were performed using a Hitachi L-6200 HPLC Pump. The following chromatographic studies were performed on an Ultimate 3000 HPLC system consisting of LPG-3400RS HPLC pump, WPS-3000TRS autosampler, TCC-3000RS column oven compartment, and VWD-3000 UV detector from Dionex (Germering, Germany). Preparation of rSPA silica monoliths
Preparation of rSPA Silica Monoliths
For this work a research prototype of Chromolith® WP 300 Epoxy was used, prepared following published processes.27, 28, 22
The immobilization of rSPA on wide pore epoxy- modified monoliths was done according to a dynamic process circulating ligand solution through the column. The rSPA ligand solution was dissolved in 6 mL immobilization buffer (50 mM sodium phosphate + 1.9 M ammonium sulfate pH 8.0) resulting in a rSPA concentration of 2 mg/mL. Before immobilization, columns were equilibrated with 50 mL immobilization buffer. The protein solution was circulated at a flow rate of 0.2 mL/min for 4 hours. Finally, the rSPA silica monolith was washed with 50 mL of 100 mM sodium phosphate buffer pH 7.4 and remaining epoxy functions were hydrolyzed with 150 mM phosphoric acid pH 1.5.
Results and Discussion
General Column Evaluation
Before the immobilization process, the silica monoliths were characterized by mercury intrusion porosimetry and nitrogen adsorption/desorption (BET). They consisted of macropores of 1.83 µm and mesopores of 28.6 nm (286 Å), resulting in a total surface area of 120 m2/g silica. The final column possessed a dynamic binding capacity of 2.75 (±0.11) mg for monoclonal IgG.
The rSPA modified silica monoliths were used for separation of monoclonal antibodies from their related impurities. The separation of cetuximab from BSA impurity was performed in just over one minute and a typical chromatogram is shown in Table 1. The unbound BSA eluted first followed by cetuximab, which was released due to the pH shift to pH 2.5. The rSPA silica monoliths provided sharp peaks with peak widths < 0.040 minutes in a short run time which was supported by the hydrophilic column surface reducing undesirable backbone interactions with the analytes.
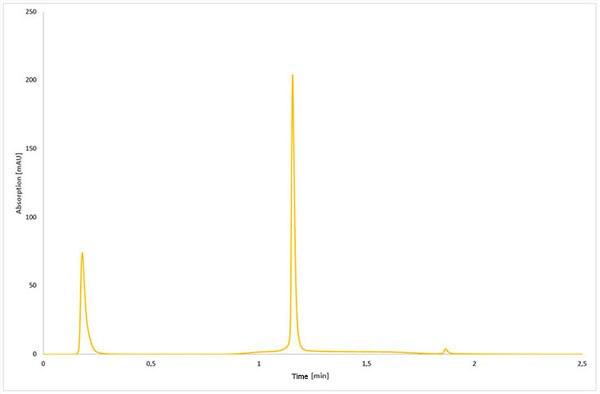
Figure 1.
Reproducibility
The cetuximab sample was injected 50 times and all chromatographic parameters were evaluated, especially with respect to their relative standard deviation (RSD). An overlay of all 50 chromatograms is shown in Figure 2 indicating a robust and reproducible column performance. The RSD of the retention time of eluted cetuximab was <0.1% with the peak width being equal for every run. The peak area RSD was 0.4%, supporting the previous data and assumptions of a robust column modification and reproducible column performance.
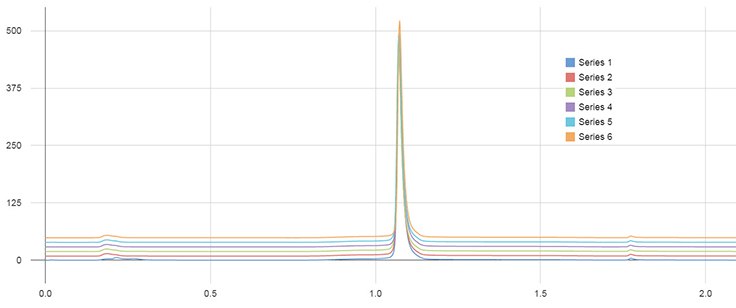
Figure 2.Analysis of 50 injections of cetuximab (1 mg/mL) on immobilized rSPA silica monolith. Only the first and every 10th chromatogram are shown. For chromatographic conditions, see Figure 1.
Furthermore, different batches were synthesized under exactly the same conditions and six columns of each batch were immobilized with rSPA ligand, respectively. Summaries of chromatographic data of each batch are shown in Table 2. The data revealed only slight differences between columns from the same batch as well as minor differences between different batches. Those results demonstrate that the immobilization process via epoxide functions is very robust and reproducible.
Column Linear Range
For titer determination, the linear range of the analytical column is a key property for its application. In antibody production, the bioreactors used contain antibody concentrations between 0.5-7 g/L, which is the minimum requirement for an analytical protein A column. The rSPA modified monolith was evaluated by analyzing different concentrations of pure cetuximab ranging from 0.0125 to 10 mg/mL. The fifteen calibration standards used covered a broad analytical range from 1.25-200 µg of injected cetuximab onto the immobilized rSPA silica monolith. The retention time of eluted cetuximab concentrations varied by only 0.1% and was confirmed by an overlay of all calibration chromatograms as shown in Figure 3. The correlation coefficient was higher than 0.999, supporting the hypothesis of a high column capacity.
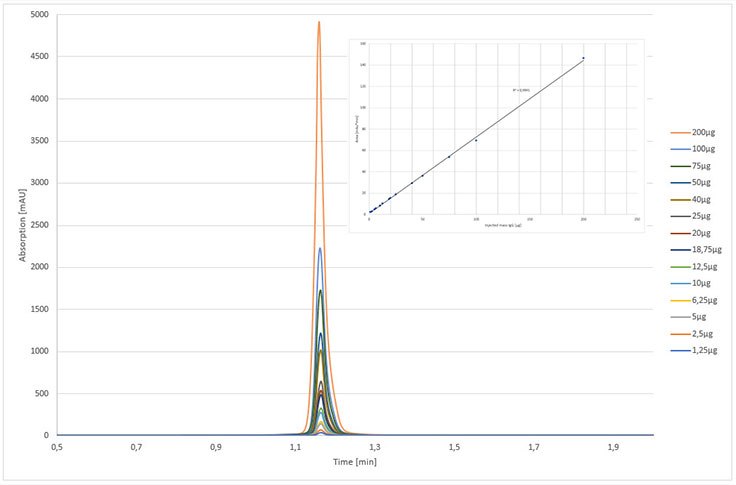
Figure 3.Calibration curve of Cetuximab on rSPA silica monolith ranging from 1.25 µg to 200 µg. Chromatographic conditions, same as in Figure 1, except injection volume: 20 µL.
Flow rate
Since monolithic silica columns exhibit a low column backpressure due to their high flow through pores, the separation capability of immobilized rSPA silica monoliths was evaluated at higher flow rates. The comparison of column performance was done with pure cetuximab spiked with BSA. It was expected that retention time and peak width of eluted cetuximab would decrease with increasing flow rate.
In Table 3, key data is shown for a flow rate comparison. It was evident that cetuximab column binding was not affected by flow rate and the relative peak area of eluted cetuximab was constant.
Additionally, column backpressure increased linearly (R2 = 0.998) enabling high flow rates and even shorter run times with constant column performance and without any loss in binding efficiency. The high-speed separation of monoclonal antibodies at high flow rates is enabled by the high mass transfer properties of Table 3. Chromatographic data of separated IgG (1 mg/mL) and BSA (1 mg/mL) by immobilized rSPA silica monolith at different flow rates. For chromatographic conditions, see Figure 1; same gradient was used as shown in Figure 1 and was adjusted based on flow rate used; flow rate: 1.0-5.0 mL/min the macropores. The flow rate data demonstrated the significant time savings and high separation efficiencies obtained by using silica monoliths.
Stability
The examination of column long-term stability was performed with IgG as a control. The stability of the monolithic structure, column modification and protein A linkage were extensively investigated by more than 5,000 runs, including 10,000 pH shifts which corresponded to more than 53,000 CV applied to the column.
The results of IgG control samples are shown in Table 4. Immobilized rSPA silica monolith provided constant analysis of IgG during the complete stability test - remaining retention time, peak area, and peak width were nearly unchanged. The RSD of IgG retention time was smaller than 0.5% whereas for the peak area of eluted IgG, the RSD was below 1.1%, indicating no loss of binding capacity.
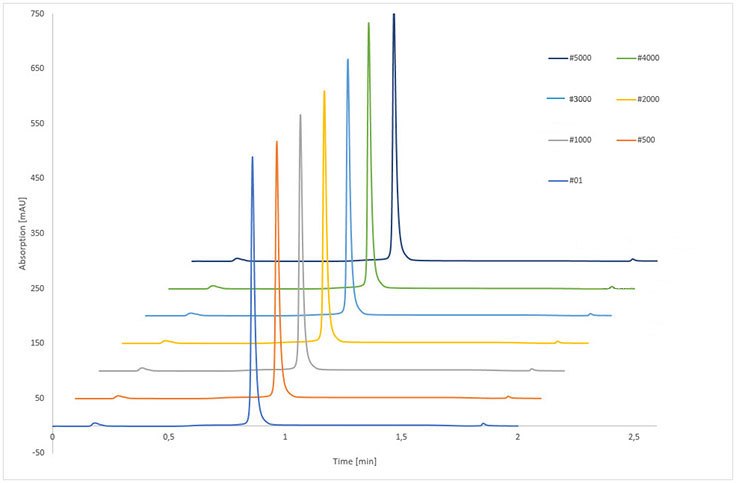
Figure 4.Stability test of immobilized rSPA silica monolith
Conclusions
Recombinant protein A was covalently attached to the surface of silica monoliths with larger mesopores suited for biomolecules. The immobilized rSPA column was successfully utilized in the separation of monoclonal antibodies by affinity chromatography. The applied method for immobilization yielded a high dynamic binding capacity, leading to a broad range of applicable antibody concentrations. Furthermore, the stability test revealed a strong linkage between silica monolith and rSPA ligand due to the constant analysis of monoclonal antibodies. In conclusion, silica monoliths with bimodal pore structure immobilized with rSPA ligand are ideally suited for the chromatographic separation of monoclonal antibodies.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?