GenElute™ Mammalian Genomic DNA Miniprep Kit Protocol ;
Product Description
The GenElute™ Mammalian Genomic DNA Miniprep Kit provides a simple and convenient way to isolate pure genomic DNA from a variety of cultured cells, tissues (including rodent tails), and fresh whole blood or white blood cells. The kit combines the advantages of silica binding with a microspin format and eliminates the need for expensive resins, alcohol precipitation, and hazardous organic compounds such as phenol and chloroform. The starting materials are lysed in a chaotropic salt-containing solution to insure the thorough denaturation of macromolecules. Addition of ethanol causes DNA to bind when the lysate is spun through a silica membrane in a microcentrifuge tube. After washing to remove contaminants, DNA is eluted in 200 µL of a Tris-EDTA solution.
Expected yields of genomic DNA will vary depending on the amount and type of starting material used (for example, 15–25 µg of RNase A-treated DNA can be isolated from 2 x 106 HeLa cells in less than one hour). DNA purified with the kit has an A260/A280 ratio between 1.6 and 1.9 and can be up to 50 kb in length. This DNA is ready for downstream applications such as restriction endonuclease digestions, PCR, Southern blots, and sequencing reactions
Reagents and Equipment Required but Not Provided
· 55 °C water bath or shaking water bath
· 70 °C water bath or heating block
· Pipette tips with aerosol barrier (recommended)
· 1.5 mL microcentrifuge tube for lysis
· Microcentrifuge (2 mL tube, rotor-equipped)*
· Ethanol (95–100%), Catalog Nos. E7148, E7023, or 459836
· Molecular Biology Reagent Water, Catalog No. W4502
*Note: To ensure proper fit of all tubes, a 24-place rotor is recommended. If you are using a
36-place rotor, we recommend using every other place for proper tube fit
Storage and Stability
Store the kit at room temperature. If any kit reagent forms a precipitate, warm at 55–65 °C until the precipitate dissolves and allow to cool to room temperature before use.
Preparation Instructions
Before beginning the procedure, do the following:
1. Preheat a water bath or shaking water bath to 55 °C
For use with tissues, rodent tails, fresh whole blood and white blood cells.
2. Preheat a water bath or heating block to 70 °C
For use with cultured cells and tissues.
3. Thoroughly mix reagents
Examine reagents for precipitation. If any reagent forms a precipitate, warm at 55–65 °C until the precipitate dissolves and allow to cool to room temperature before use.
4. Dilute Wash Solution Concentrate
Dilute the concentrate with 10 mL (10 prep package), 80 mL (70 prep package) or 360 mL (350 prep package) of 95–100% ethanol. After each use, tightly cap the diluted Wash Solution to prevent the evaporation of ethanol.
5. Dissolve Proteinase K in water
According to Table 1 below, dissolve the powder in one bottle of Proteinase K in water (Catalog No. W4502) to obtain a 20 mg/mL stock solution. This solution can be stored for several days at 2–8 °C. For longer-term storage, the unused portion of the solution may be stored in aliquots at –20 °C until needed. This product as supplied is stable at room temperature.
The Proteinase K solution must be added directly to each sample preparation every time. Do not combine the Proteinase K and Lysis Solutions for storage.
Procedure
Note: If minimally sheared genomic DNA is desired in downstream applications, e.g., if using the end product for long amplification PCR, mix with gentle pipetting or inversion until homogeneous instead of vortexing in the procedure that follows.
A. Cultured Cell Preparation
1a. Harvest cells
• Attached cell cultures: Release cells with trypsin. Pellet up to 5 × 106 cells for 5 minutes at 300 × g; remove the culture medium completely and discard. Continue to step 2a.
• Suspension cell cultures: Pellet up to 5 × 106 cells for 5 minutes at 300 × g; remove the culture medium completely and discard. Continue with step 2a.
Note: Cells can be harvested, aliquotted into 1.5 mL microcentrifuge tubes and flash-frozen in liquid nitrogen, then stored at
–70 °C for several months before preparing DNA.
2a. Resuspend cells
Resuspend the pellet thoroughly in 200 µL of Resuspension Solution. If previously frozen, allow the cell pellet to thaw slightly before resuspending. If residual RNA is not a concern, continue with step 3a.
Optional RNase A treatment: If RNA-free genomic DNA is required, add 20 µL of RNase A Solution and incubate for 2 minutes at room temperature, then continue with step 3a.
3a. Lyse cells
Add 20 µL of the Proteinase K solution to the sample, followed by 200 µL of Lysis Solution C (B8803). Vortex thoroughly (about 15 seconds), and incubate at 70 °C for 10 minutes. A homogeneous mixture is essential for efficient lysis. Continue with step 4.
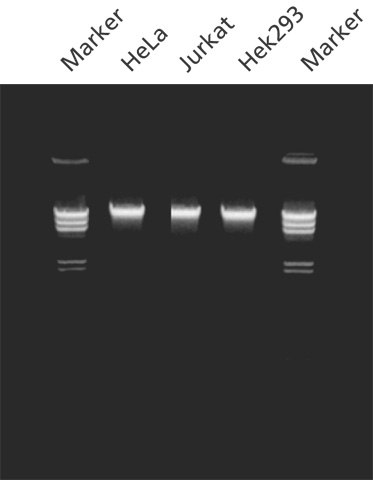
Figure 1. Genomic DNA purified from cells using GenElute™ Mammalian Genomic DNA Purification Kit. Purified genomic DNA was isolated with the GenElute™ Mammalian Genomic DNA Purification Kit from 2 x 106 cells of the sources indicated. The genomic DNA (200 ng/lane) was analyzed on a 0.8% agarose gel. Markers are lambda DNA digested with Hind III.
B. Mammalian Tissue Preparation
1b. Prepare tissue
Quickly mince and weigh a piece of fresh or frozen tissue. Allow frozen tissue to thaw slightly before slicing, but keep on ice to protect against degradation. Cutting tissue into small pieces enables more efficient lysis. Up to 25 mg of tissue (or 10 mg of spleen, due to the high number of cells per given mass) may be used per preparation. Transfer to a 1.5 mL microcentrifuge tube and continue to step 2b.
Note: Tissue can be harvested, aliquotted into 1.5 mL microcentrifuge tubes and flash-frozen in liquid nitrogen, then stored at
–70 °C for several months before preparing the DNA.
2b. Digest tissue
Add 180 µl of Lysis Solution T (B6678), followed by 20 µL of the Proteinase K solution to the tissue. Mix by vortexing. Incubate the sample at 55 °C until the tissue is completely digested and no particles remain. Vortex occasionally or use a shaking water bath. Digestion is usually complete in 2 to 4 hours. After digestion is complete, vortex briefly.
Optional RNase A treatment: If residual RNA is not a concern, continue with step 3b. If RNA-free genomic DNA is required, add 20 µL of RNase A Solution and incubate for 2 minutes at room temperature, then continue with step 3b.
3b. Lyse cells
Add 200 µL of Lysis Solution C (B8803) to the sample; vortex thoroughly (15 seconds). A homogeneous mixture is essential for efficient lysis. Incubate at 70 °C for 10 minutes. Continue with step 4.

Figure 2. Genomic DNA purified from tissues using GenElute™ Mammalian Genomic DNA Purification Kit. Purified genomic DNA, from the indicated mouse tissues, was isolated with the GenElute™ Mammalian Genomic DNA Purification Kit. Genomic DNA (200 ng/lane) was analyzed on a 0.8% agarose gel to illustrate yield and integrity. Markers are lambda DNA digested with Hind III.
C. Rodent Tail Preparation
1c. Prepare rodent tails
Quickly measure and cut a piece of fresh or frozen rodent tail. Allow frozen rodent tail to thaw slightly before cutting, but keep on ice to protect against degradation. Do not use more than 0.6 cm (rat) or 1.2 cm (mouse) tail per preparation. Cut two (mouse) or one (rat) 0.5–0.6 cm lengths of tail and place them in a 1.5 mL microcentrifuge tube; continue to step 2c.
Note: Rodent tails can be stored at –20 °C for several months before preparing DNA.
2c. Digest tissue
Add 180 µL of Lysis Solution T (B6678), followed by 20 µL of the Proteinase K solution to the rodent tail. Mix by vortexing; ensure that the tail is fully submerged. Incubate the sample at 55 °C until the tail tissue is completely digested. Some particles (bone and hair) may remain. Vortex occasionally or use a shaking water bath during incubation for more rapid digestion. Digestion is usually complete in 3 to 6 hours. After digestion is complete, vortex briefly. If residual RNA is not a concern, continue with step 3.
Optional RNase A treatment: If RNA-free genomic DNA is required, add 20 µL of RNase A Solution and incubate for 2 minutes at room temperature; continue with step 3c.
3c. Lyse cells
Add 200 µL of Lysis Solution C (B8803) to the sample; vortex thoroughly (15 seconds). A homogeneous mixture is essential for efficient lysis. Continue with step 4.
D. Whole Blood Preparation
1d. Collect blood
Collect whole blood in an anticoagulant tube (an EDTA tube is preferred). Whole blood should be equilibrated to room temperature before beginning the preparation.
2d. Prepare blood
Place 20 µL of the Proteinase K solution into a 1.5 µL microcentrifuge tube. Add up to 200 µL of the whole blood sample to the tube. If the sample is less than 200 µL, add the Resuspension Solution to bring the volume up to 200 µL.
Note: If the sample is already dispensed in a tube, the Proteinase K solution can be added to the sample. Vortex to ensure thorough mixing of the enzyme. Whole blood can be stored at 4 °C for several hours before preparing DNA. If residual RNA is not a concern, continue with step 3d.
Optional RNase A treatment: If RNA-free genomic DNA is required, add 20 µL of RNase A Solution and incubate for 2 minutes at room temperature; continue with step 3d.
3d. Lyse cells
Add 200 µL of Lysis Solution C (B8803) to the sample; vortex thoroughly (15 seconds). A homogeneous mixture is essential for efficient lysis. Incubate at 55 °C for 10 minutes. Continue with step 4.
E. White Blood Cell (WBC) Preparation
1e. Prepare white blood cells from whole blood
Prepare WBCs from 500 µL of whole blood per preparation; Appendix for an ammonium chloride lysis procedure.
2e. Resuspend cells
Resuspend the pellet thoroughly in 200 µL of the Resuspension Solution; add 20 µL of the Proteinase K solution to the sample and vortex briefly to ensure thorough mixing of the enzyme. If residual RNA is not a concern, continue with step 3e.
Optional RNase A treatment: If RNA-free genomic DNA is required, add 20 µL of RNase A Solution and incubate for 2 minutes at room temperature, then continue with step 3e.
3e. Lyse cells
Add 200 µl of Lysis Solution C (B8803) to the sample; vortex thoroughly. A homogeneous mixture is essential for efficient lysis. Incubate at 55 °C for 10 minutes. Continue with step 4.
DNA Isolation From All Listed Sample Types
This is a continuation of the procedure from the samples prepared in Sections A, B, C, D, and E.
4. Column preparation
Add 500 µL of the Column Preparation Solution to each pre-assembled GenElute™ Miniprep Binding Column (with a red o-ring, not to be confused with other GenElute™ kits) and centrifuge at 12,000 × g for 1 minute. Discard flowthrough liquid.
Note: The Column Preparation Solution maximizes binding of DNA to the membrane resulting in more consistent yields.
5. Prepare for binding
Add 200 µL of ethanol (95–100%) to the lysate; mix thoroughly by vortexing 5–10 seconds. A homogeneous solution is essential.
6. Load lysate
Transfer the entire contents of the tube into the treated binding column from step 4. Use a wide bore pipette tip to reduce shearing the DNA when transferring contents into the binding column. Centrifuge at ≥6500 × g for 1 minute. Discard the collection tube containing the flowthrough liquid and place the binding column in a new 2 mL collection tube.
7. First wash
Prior to first use, dilute the Wash Solution Concentrate with ethanol as described under Preparation Instructions. Add 500 µL of Wash Solution to the binding column and centrifuge for 1 minute at ≥6,500 × g. Discard the collection tube containing the flow-through liquid and place the binding column in a new 2 mL collection tube.
8. Second wash
Add another 500 µL of Wash Solution to the binding column; centrifuge for 3 minutes at maximum speed (12,000-16,000 × g) to dry the binding column. The binding column must be free of ethanol before eluting the DNA. Centrifuge the column for one additional minute at maximum speed if residual ethanol is seen. You may empty and re-use the collection tube if you need this additional centrifugation step. Finally, discard the collection tube containing the flowthrough liquid and place the binding column in a new 2 mL collection tube.
9. Elute DNA
Pipette 200 µL of the Elution Solution directly into the center of the binding column; centrifuge for 1 minute at ≥6,500 × g to elute the DNA. To increase the elution efficiency, incubate for 5 minutes at room temperature after adding the Elution Solution, then centrifuge.
Optional: A second elution can be collected by repeating step 9 with an additional 200 µL of Elution Solution and eluting into a new 2 mL collection tube (provided) or into the same 2 mL collection tube as used for the first eluate.
The eluate contains pure genomic DNA. For short term storage of DNA, 2–8 °C is recommended. For long-term storage of DNA, -20 °C is recommended. Avoid freezing and thawing, which causes breaks in the DNA strand. The Elution Solution will help stabilize the DNA at these temperatures.
DNA Precipitation (Optional)
The GenElute™ Mammalian Genomic DNA Kit is designed so that the DNA always remains in solution, thus avoiding resuspension issues. However, if you find it necessary to concentrate the DNA, ethanol precipitation in the presence of sodium acetate is recommended.
Results
The concentration and quality of the genomic DNA prepared with the GenElute™ kit can be determined by spectrophotometric analysis and agarose gel electrophoresis. Dilute the DNA in TE Buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0–8.5) and measure the absorbance at 260 nm and 280 nm using a quartz microcuvette. The absorbance should be between 0.1 and 1.0 (or within the linear range of your spectrophotometer). An absorbance of 1.0 at 260 nm corresponds to approximately 50 µg/mL of doublestranded DNA. The ratio of absorbance at 260 nm to 280 nm (A260/A280) should be 1.6–1.9. The size and quality of the DNA can be determined by agarose gel electrophoresis.1 A gel containing 0.8% agarose (Catalog No. A9539) in 0.5X TBE Buffer (Catalog No. T6400) works well for the resolution of genomic DNA. The DNA can be visualized by staining with an intercalating dye such as ethidium bromide (Catalog No. E1510) and measured against a known DNA marker such as Lambda DNA EcoRI Hind III digest (Catalog No. D9281). The genomic DNA should migrate as a single, high molecular weight band with very little evidence of shearing. A more precise determination of the size of the DNA can be made by pulsed-field gel electrophoresis.
Troubleshooting Guide
1. Binding column is clogged
Cause — Sample is too large.
Solution — In the future, use fewer cells, smaller pieces of tissue, smaller pieces of rodent tail, or a smaller quantity of whole blood. To salvage the current preparation, increase the g-force and/or spin longer until lysate passes through the binding column. The yield of genomic DNA may be reduced.
Cause — Tissue or rodent tail is inefficiently disrupted.
Solution — Extend the Proteinase K digestion at 55 °C. To expedite lysis, cut tissue or rodent tail into smaller pieces and mix frequently during digestion to ensure more efficient lysis. Inverting the sample tube after Proteinase K digestion will ensure a more homogeneous mixture. Do not use a combined solution (Proteinase K + Lysis Solution) that has been stored.
2. Low yield of genomic DNA
Cause — Sample may be old or degraded OR Tissue is inefficiently disrupted.
Solution — The yield will vary among different types of cells, tissues, and fresh whole blood. Use cultures before they reach maximum density or as they become fully confluent. Harvest tissues or rodent tails as rapidly as possible. Use whole blood within a few hours. If samples are being stored for future use, flash-freeze the cells or tissue in liquid nitrogen. Whole blood should be kept at 4 °C no longer than 12 hours.
Cause — Lysate/ethanol mixture is not homogeneous.
Solution — To ensure a homogeneous solution, vortex 5–10 seconds before applying to the binding column. If minimally sheared genomic DNA is desired in downstream applications, e.g. if using the end product for long amplification PCR, mix with gentle pipetting or inversion until homogeneous instead of vortexing.
Cause — Lysate/ethanol mixture is not homogeneous.
Solution — To ensure a homogeneous solution, vortex 5–10 seconds before applying to the binding column. If minimally sheared genomic DNA is desired in downstream applications, e.g. if using the end product for long amplification PCR, mix with gentle pipetting or inversion until homogeneous instead of vortexing.
Cause — DNA elution is incomplete.
Solution — Confirm that the DNA was eluted in 200 µL of Elution Solution. The DNA yield for most types of material may be improved by incubating the Elution Solution for 5 minutes at room temperature after it is added to the column. A second and third elution using 200 µL of Elution Solution may be performed.
Cause — Ethanol was omitted during binding.
Solution — Check that the ethanol was added in step 5 before applying the sample to the binding column in step 6.
Cause — The eluate contains residual ethanol from the wash solution.
Solution — Ethanol from the final wash must be eliminated before eluting the DNA. Spin longer or a second time to dry the membrane. If flow-through liquid containing ethanol contacts the binding column, repeat the centrifugation step before eluting the DNA.
Cause — The eluate contains residual ethanol from the wash solution.
Solution — Ethanol from the final wash must be eliminated before eluting the DNA. Spin longer or a second time to dry the membrane. If flow-through liquid containing ethanol contacts the binding column, repeat the centrifugation step before eluting the DNA.
Cause — The eluate contains residual ethanol from the wash solution.
Solution — Ethanol from the final wash must be eliminated before eluting the DNA. Spin longer or a second time to dry the membrane. If flow-through liquid containing ethanol contacts the binding column, repeat the centrifugation step before eluting the DNA.
Cause — Wash Solution Concentrate was not diluted before use.
Solution — Check that the Wash Solution Concentrate was properly diluted with ethanol before use.
Cause — Wash Solution Concentrate was not diluted before use.
Solution — Check that the Wash Solution Concentrate was properly diluted with ethanol before use.
Cause — DNA was eluted with water instead of Elution Solution.
Solution — Elution Solution is recommended for optimal yields and storage of the purified DNA. If water is used to elute the DNA, confirm that the pH is at least 7.0, to avoid acidic conditions which may subject the DNA to acid hydrolysis when stored for long periods of time.
3. Purity of the DNA is lower than expected; A260/A280 ratio is too low
Cause — Sample was diluted in water.
Solution — Use either Elution Solution (10 mM Tris-HCl, 0.5 mM EDTA, pH 9.0) or 10 mM Tris-HCl, pH 8.0–8.5 as the eluant.
Cause — Background reading is high due to silica fines.
Solution — Spin the DNA sample at maximum speed for 1 minute; use the supernatant to repeat the absorbance readings.
4. Purity of the DNA is lower than expected; A260/A280 ratio is too high
Cause — Genomic DNA is contaminated with RNA.
Solution — Include an RNase A treatment step in future isolations or treat final product with RNase A Solution and repurify.
5. DNA is sheared
Cause — Genomic DNA was handled improperly.
Solution — This kit was designed to eliminate DNA precipitation and common resuspension steps found in other genomic DNA kits that can lead to shearing. All pipetting steps should be executed as gently as possible. Wide-orifice pipette tips are recommended to help eliminate shearing. If minimally sheared genomic DNA is desired in downstream applications, e.g., if using the end product for long amplification PCR, mix with gentle pipetting or inversion until homogeneous instead of vortexing.
Cause — Sample is old, degraded, or has undergone repeated freeze/thaw cycles.
Solution — Old starting material may yield degraded DNA in the eluate. Fresh cell and tissue, rodent tail, and whole blood preparations should be used immediately. Alternatively, cells and tissue can be frozen in liquid nitrogen and stored at –70 °C until needed. Rodent tails can be stored at –20 °C for several weeks or –70 °C for several months. Whole blood can be stored at 4 °C for up to 12 hours.
6. Downstream applications are inhibited
Cause — Ethanol is carried over into the final genomic DNA preparation.
Solution — After the final wash of the binding column (step 8), do not allow the flow-through liquid to contact the column. Re-spin the column, if necessary, after emptying the collection tube, for an additional 1 minute at maximum speed (12,000–16,000 × g).
Cause — Salt is carried over into the final genomic DNA preparation.
Solution — Make sure that the binding column is transferred to a new 2 mL collection tube before adding the Wash Solution in step 8.
Appendix: Ammonium Chloride Lysis Procedure for Isolating White Blood Cells from Whole Blood
1. Collect whole blood in an anticoagulant tube.
2. Add 500 µL of whole blood to 1 mL of ammonium chloride lysis solution*. Gently mix (by inversion or on a table rocker) at room temperature for 5 minutes, then spin for 5 minutes at 700 × g in a microcentrifuge.
3. Discard supernatant. Gently resuspend pellet in one additional mL of ammonium chloride lysis solution; mix gently and spin as in step 2. Discard the supernatant, keep the cell pellet on ice and resume the White Blood Cell Protocol (1e).
*Ammonium Chloride Lysis Solution:
160 mM ammonium chloride (Catalog No. A9434), 20 mM sodium bicarbonate (Catalog No. S7277); pH to 7.2 with HCl.
Materials
Precautions and Disclaimer
The GenElute™ Mammalian Genomic DNA Kit is for laboratory use only, not for drug, household, or other uses. The Lysis Solution C contains a chaotropic salt, which is an irritant. The Column Preparation Solution is an irritant. Avoid contact with skin. Wear gloves, safety glasses, and suitable protective clothing when handling these solutions or any reagent provided with the kit. Please consult the Safety Data Sheet (SDS) for information regarding hazards and safe handling practices.
References
To continue reading please sign in or create an account.
Don't Have An Account?